Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium
- PMID: 30484918
- PMCID: PMC6417950
- DOI: 10.1111/mmi.14174
Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium
Abstract
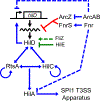
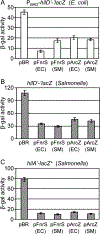
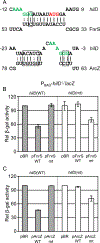
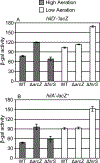
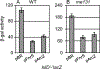
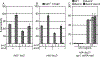
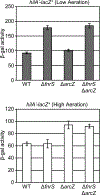
Salmonella Typhimurium induces inflammatory diarrhea and uptake into intestinal epithelial cells using the Salmonella pathogenicity island 1 (SPI1) type III secretion system (T3SS). Three AraC-like regulators, HilD, HilC and RtsA, form a feed-forward regulatory loop that activates transcription of hilA, encoding the activator of the T3SS structural genes. Many environmental signals and regulatory systems are integrated into this circuit to precisely regulate SPI1 expression. A subset of these regulatory factors affects translation of hilD, but the mechanisms are poorly understood. Here, we identified two sRNAs, FnrS and ArcZ, which repress hilD translation, leading to decreased production of HilA. FnrS and ArcZ are oppositely regulated in response to oxygen, one of the key environmental signals affecting expression of SPI1. Mutational analysis demonstrates that FnrS and ArcZ bind to the hilD mRNA 5' UTR, resulting in translational repression. Deletion of fnrS led to increased HilD production under low-aeration conditions, whereas deletion of arcZ abolished the regulatory effect on hilD translation aerobically. The fnrS arcZ double mutant has phenotypes in a mouse oral infection model consistent with increased expression of SPI1. Together, these results suggest that coordinated regulation by these two sRNAs maximizes HilD production at an intermediate level of oxygen.
© 2018 John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Figures




References
-
- (2005) Northern blotting: transfer of denatured RNA to membranes. Nature Methods 2: 997.
-
- Ares M (2012) Bacterial RNA Isolation. Cold Spring Harbor Protocols 2012: pdb.prot071068. - PubMed
-
- Argenzio RA, Southworth M and Stevens CE (1974) Sites of organic acid production and absorption in the equine gastrointestinal tract. Am J Physiol 226: 1043–1050. - PubMed
-
- Bajaj V, Lucas RL, Hwang C and Lee CA (1996) Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22: 703–714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

