Identification of a 4-mRNA metastasis-related prognostic signature for patients with breast cancer
- PMID: 30484951
- PMCID: PMC6349190
- DOI: 10.1111/jcmm.14049
Identification of a 4-mRNA metastasis-related prognostic signature for patients with breast cancer
Abstract
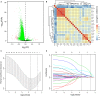
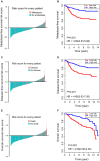
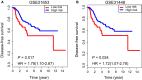
Metastasis-related mRNAs have showed great promise as prognostic biomarkers in various types of cancers. Therefore, we attempted to develop a metastasis-associated gene signature to enhance prognostic prediction of breast cancer (BC) based on gene expression profiling. We firstly screened and identified 56 differentially expressed mRNAs by analysing BC tumour tissues with and without metastasis in the discovery cohort (GSE102484, n = 683). We then found 26 of these differentially expressed genes were associated with metastasis-free survival (MFS) in the training set (GSE20685, n = 319). A metastasis-associated gene signature built using a LASSO Cox regression model, which consisted of four mRNAs, can classify patients into high- and low-risk groups in the training cohort. Patients with high-risk scores in the training cohort had shorter MFS (hazard ratio [HR] 3.89, 95% CI 2.53-5.98; P < 0.001), disease-free survival (DFS) (HR 4.69, 2.93-7.50; P < 0.001) and overall survival (HR 4.06, 2.56-6.45; P < 0.001) than patients with low-risk scores. The prognostic accuracy of mRNAs signature was validated in the two independent validation cohorts (GSE21653, n = 248; GSE31448, n = 246). We then developed a nomogram based on the mRNAs signature and clinical-related risk factors (T stage and N stage) that predicted an individual's risk of disease, which can be assessed by calibration curves. Our study demonstrated that this 4-mRNA signature might be a reliable and useful prognostic tool for DFS evaluation and will facilitate tailored therapy for BC patients at different risk of disease.
Keywords: breast cancer; mRNAs signature; prognosis; sisease-free survival.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures


Similar articles
-
Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study.Lancet Oncol. 2018 Mar;19(3):382-393. doi: 10.1016/S1470-2045(18)30080-9. Epub 2018 Feb 7. Lancet Oncol. 2018. PMID: 29428165
-
A 16-mRNA signature optimizes recurrence-free survival prediction of Stages II and III gastric cancer.J Cell Physiol. 2020 Jul;235(7-8):5777-5786. doi: 10.1002/jcp.29511. Epub 2020 Feb 11. J Cell Physiol. 2020. PMID: 32048287
-
Recurrence-associated gene signature optimizes recurrence-free survival prediction of colorectal cancer.Mol Oncol. 2017 Nov;11(11):1544-1560. doi: 10.1002/1878-0261.12117. Epub 2017 Sep 23. Mol Oncol. 2017. PMID: 28796930 Free PMC article.
-
Identification and validation of a potent multi-mRNA signature for the prediction of early relapse in hepatocellular carcinoma.Carcinogenesis. 2019 Jul 20;40(7):840-852. doi: 10.1093/carcin/bgz018. Carcinogenesis. 2019. PMID: 31059567
-
Identification of a New Eight-Long Noncoding RNA Molecular Signature for Breast Cancer Survival Prediction.DNA Cell Biol. 2019 Dec;38(12):1529-1539. doi: 10.1089/dna.2019.5059. Epub 2019 Oct 24. DNA Cell Biol. 2019. PMID: 31647329
Cited by
-
CircKIF4A promotes glioma growth and temozolomide resistance by accelerating glycolysis.Cell Death Dis. 2022 Aug 27;13(8):740. doi: 10.1038/s41419-022-05175-z. Cell Death Dis. 2022. PMID: 36030248 Free PMC article.
-
The Pan-Cancer Multi-Omics Landscape of FOXO Family Relevant to Clinical Outcome and Drug Resistance.Int J Mol Sci. 2022 Dec 9;23(24):15647. doi: 10.3390/ijms232415647. Int J Mol Sci. 2022. PMID: 36555288 Free PMC article.
-
circEPSTI1 promotes tumor progression and cisplatin resistance via upregulating MSH2 in cervical cancer.Aging (Albany NY). 2022 Jul 2;14(13):5406-5416. doi: 10.18632/aging.204152. Epub 2022 Jul 2. Aging (Albany NY). 2022. PMID: 35779530 Free PMC article.
-
Comprehensive analysis of glycolysis mediated pattern clusters and immune infiltration characterization of tumor microenvironment in triple-negative breast cancer.Heliyon. 2023 Apr 1;9(4):e15175. doi: 10.1016/j.heliyon.2023.e15175. eCollection 2023 Apr. Heliyon. 2023. PMID: 37089355 Free PMC article.
-
Identification of a prognostic 4-mRNA signature in laryngeal squamous cell carcinoma.J Cancer. 2021 Aug 3;12(19):5807-5816. doi: 10.7150/jca.47557. eCollection 2021. J Cancer. 2021. PMID: 34475994 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7‐30. - PubMed
-
- DeSantis CE, Ma J, Goding SA, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439‐448. - PubMed
-
- Wang M, Chen H, Wu K, et al. Evaluation of the prognostic stage in the 8th edition of the American Joint Committee on Cancer in locally advanced breast cancer: an analysis based on SEER 18 database. Breast. 2018;37:56‐63. - PubMed
-
- Kim JY, Lim JE, Jung HH, et al. Validation of the new AJCC eighth edition of the TNM classification for breast cancer with a single‐center breast cancer cohort. Breast Cancer Res Treat. 2018;171:737. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

