Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance
- PMID: 30485383
- PMCID: PMC6287881
- DOI: 10.1371/journal.ppat.1007420
Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance
Abstract
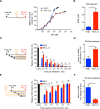
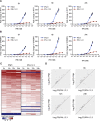
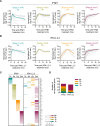
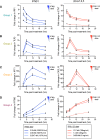
It is currently believed that type I and III interferons (IFNs) have redundant functions. However, the preferential distribution of type III IFN receptor on epithelial cells suggests functional differences at epithelial surfaces. Here, using human intestinal epithelial cells we could show that although both type I and type III IFNs confer an antiviral state to the cells, they do so with distinct kinetics. Type I IFN signaling is characterized by an acute strong induction of interferon stimulated genes (ISGs) and confers fast antiviral protection. On the contrary, the slow acting type III IFN mediated antiviral protection is characterized by a weaker induction of ISGs in a delayed manner compared to type I IFN. Moreover, while transcript profiling revealed that both IFNs induced a similar set of ISGs, their temporal expression strictly depended on the IFNs, thereby leading to unique antiviral environments. Using a combination of data-driven mathematical modeling and experimental validation, we addressed the molecular reason for this differential kinetic of ISG expression. We could demonstrate that these kinetic differences are intrinsic to each signaling pathway and not due to different expression levels of the corresponding IFN receptors. We report that type III IFN is specifically tailored to act in specific cell types not only due to the restriction of its receptor but also by providing target cells with a distinct antiviral environment compared to type I IFN. We propose that this specific environment is key at surfaces that are often challenged with the extracellular environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
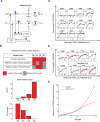
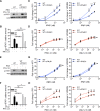
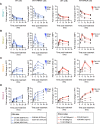
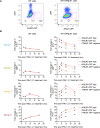
References
-
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131–7. 10.1038/ni1303 - DOI - PubMed
-
- Koyama S, Ishii KJ, Coban C, Akira S. Innate immune response to viral infection. Cytokine. 2008;43(3):336–41. 10.1016/j.cyto.2008.07.009 - DOI - PubMed
-
- Kotenko S V, Gallagher G, Baurin V V, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. 10.1038/ni875 - DOI - PubMed
-
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–8. 10.1038/ni873 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

