Potential limits to the benefits of admixture during biological invasion
- PMID: 30485593
- PMCID: PMC6344275
- DOI: 10.1111/mec.14958
Potential limits to the benefits of admixture during biological invasion
Abstract
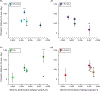
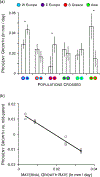
Species introductions often bring together genetically divergent source populations, resulting in genetic admixture. This geographic reshuffling of diversity has the potential to generate favourable new genetic combinations, facilitating the establishment and invasive spread of introduced populations. Observational support for the superior performance of admixed introductions has been mixed, however, and the broad importance of admixture to invasion questioned. Under most underlying mechanisms, admixture's benefits should be expected to increase with greater divergence among and lower genetic diversity within source populations, though these effects have not been quantified in invaders. We experimentally crossed source populations differing in divergence in the invasive plant Centaurea solstitialis. Crosses resulted in many positive (heterotic) interactions, but fitness benefits declined and were ultimately negative at high source divergence, with patterns suggesting cytonuclear epistasis. We explored the literature to assess whether such negative epistatic interactions might be impeding admixture at high source population divergence. Admixed introductions reported for plants came from sources with a wide range of genetic variation, but were disproportionately absent where there was high genetic divergence among native populations. We conclude that while admixture is common in species introductions and often happens under conditions expected to be beneficial to invaders, these conditions may be constrained by predictable negative genetic interactions, potentially explaining conflicting evidence for admixture's benefits to invasion.
Keywords: cytonuclear interactions; epistasis; genetic diversity; heterosis; invasiveness; multiple introductions.
© 2018 John Wiley & Sons Ltd.
Figures


Similar articles
-
Population genomic analyses reveal a history of range expansion and trait evolution across the native and invaded range of yellow starthistle (Centaurea solstitialis).Mol Ecol. 2017 Feb;26(4):1131-1147. doi: 10.1111/mec.13998. Epub 2017 Feb 4. Mol Ecol. 2017. PMID: 28029713 Free PMC article.
-
Native and Invading Yellow Starthistle (Centaurea solstitialis) Microbiomes Differ in Composition and Diversity of Bacteria.mSphere. 2019 Mar 6;4(2):e00088-19. doi: 10.1128/mSphere.00088-19. mSphere. 2019. PMID: 30842267 Free PMC article.
-
The devil is in the details: genetic variation in introduced populations and its contributions to invasion.Mol Ecol. 2015 May;24(9):2095-111. doi: 10.1111/mec.13183. Epub 2015 Apr 21. Mol Ecol. 2015. PMID: 25846825 Review.
-
Effects of admixture in native and invasive populations of Lythrum salicaria.Biol Invasions. 2018;20(9):2381-2393. doi: 10.1007/s10530-018-1707-2. Epub 2018 Mar 21. Biol Invasions. 2018. PMID: 30956538 Free PMC article.
-
How important is intraspecific genetic admixture to the success of colonising populations?Trends Ecol Evol. 2014 Apr;29(4):233-42. doi: 10.1016/j.tree.2014.02.003. Epub 2014 Mar 15. Trends Ecol Evol. 2014. PMID: 24636862 Review.
Cited by
-
Spatially Varying Wolbachia Frequencies Reveal the Invasion Origin of an Agricultural Pest Recently Introduced From Europe to North America.Evol Appl. 2024 Sep 20;17(9):e70016. doi: 10.1111/eva.70016. eCollection 2024 Sep. Evol Appl. 2024. PMID: 39310793 Free PMC article.
-
Invader at the edge - Genomic origins and physiological differences of round gobies across a steep urban salinity gradient.Evol Appl. 2022 Jul 16;16(2):321-337. doi: 10.1111/eva.13437. eCollection 2023 Feb. Evol Appl. 2022. PMID: 36793700 Free PMC article.
-
Population genomic and historical analysis suggests a global invasion by bridgehead processes in Mimulus guttatus.Commun Biol. 2021 Mar 12;4(1):327. doi: 10.1038/s42003-021-01795-x. Commun Biol. 2021. PMID: 33712659 Free PMC article.
-
Predicting species invasiveness with genomic data: Is genomic offset related to establishment probability?Evol Appl. 2024 Jun 14;17(6):e13709. doi: 10.1111/eva.13709. eCollection 2024 Jun. Evol Appl. 2024. PMID: 38884022 Free PMC article.
-
Genetic diversity pattern reveals the primary determinant of burcucumber (Sicyos angulatus L.) invasion in Korea.Front Plant Sci. 2022 Nov 15;13:997521. doi: 10.3389/fpls.2022.997521. eCollection 2022. Front Plant Sci. 2022. PMID: 36457533 Free PMC article.
References
-
- Allendorf FW, Lundquist LL (2003) Introduction: population biology, evolution, and control of invasive species. Conservation Biology, 17, 24–30.
-
- Bailey MF, McCauley DE (2006) The effects of inbreeding, outbreeding and long-distance gene flow on survivorship in North American populations of Silene vulgaris. Journal of Ecology, 94, 98–109.
-
- Baker HG, Stebbins GL (Eds.) (1965) The Genetics of Colonizing Species. Academic Press, New York.
-
- Ballard JWO, Melvin RG (2010) Linking the mitochondrial genotype to the organismal phenotype. Molecular Ecology, 19, 1523–1539. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

