Esrp1-Regulated Splicing of Arhgef11 Isoforms Is Required for Epithelial Tight Junction Integrity
- PMID: 30485810
- PMCID: PMC6371790
- DOI: 10.1016/j.celrep.2018.10.097
Esrp1-Regulated Splicing of Arhgef11 Isoforms Is Required for Epithelial Tight Junction Integrity
Abstract
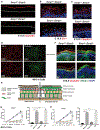
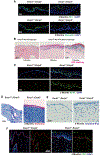
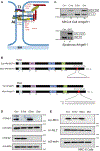
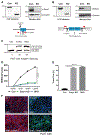
The epithelial-specific splicing regulators Esrp1 and Esrp2 are required for mammalian development, including establishment of epidermal barrier functions. However, the mechanisms by which Esrp ablation causes defects in epithelial barriers remain undefined. We determined that the ablation of Esrp1 and Esrp2 impairs epithelial tight junction (TJ) integrity through loss of the epithelial isoform of Rho GTP exchange factor Arhgef11. Arhgef11 is required for the maintenance of TJs via RhoA activation and myosin light chain (MLC) phosphorylation. Ablation or depletion of Esrp1/2 or Arhgef11 inhibits MLC phosphorylation and only the epithelial Arhgef11 isoform rescues MLC phosphorylation in Arhgef11 KO epithelial cells. Mesenchymal Arhgef11 transcripts contain a C-terminal exon that binds to PAK4 and inhibits RhoA activation byArhgef11. Deletion of the mesenchymal-specific Arhgef11 exon in Esrp1/2 KO epithelial cells using CRISPR/Cas9 restored TJ function, illustrating how splicing alterations can be mechanistically linked to disease phenotypes that result from impaired functions of splicing regulators.
Keywords: epidermis.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Rho GTP exchange factor ARHGEF11 regulates the integrity of epithelial junctions by connecting ZO-1 and RhoA-myosin II signaling.Proc Natl Acad Sci U S A. 2012 Jun 19;109(25):9905-10. doi: 10.1073/pnas.1115063109. Epub 2012 Jun 4. Proc Natl Acad Sci U S A. 2012. PMID: 22665792 Free PMC article.
-
Transcriptomic analysis of PNN- and ESRP1-regulated alternative pre-mRNA splicing in human corneal epithelial cells.Invest Ophthalmol Vis Sci. 2013 Jan 23;54(1):697-707. doi: 10.1167/iovs.12-10695. Invest Ophthalmol Vis Sci. 2013. PMID: 23299472 Free PMC article.
-
Ablation of the epithelial-specific splicing factor Esrp1 results in ureteric branching defects and reduced nephron number.Dev Dyn. 2016 Oct;245(10):991-1000. doi: 10.1002/dvdy.24431. Epub 2016 Jul 28. Dev Dyn. 2016. PMID: 27404344 Free PMC article.
-
Roles and Regulation of Epithelial Splicing Regulatory Proteins 1 and 2 in Epithelial-Mesenchymal Transition.Int Rev Cell Mol Biol. 2016;327:163-194. doi: 10.1016/bs.ircmb.2016.06.003. Epub 2016 Jul 30. Int Rev Cell Mol Biol. 2016. PMID: 27692175 Review.
-
Epithelial junctions and Rho family GTPases: the zonular signalosome.Small GTPases. 2014;5(4):1-15. doi: 10.4161/21541248.2014.973760. Small GTPases. 2014. PMID: 25483301 Free PMC article. Review.
Cited by
-
Maximizing the Utility of Transcriptomics Data in Inflammatory Skin Diseases.Front Immunol. 2021 Oct 29;12:761890. doi: 10.3389/fimmu.2021.761890. eCollection 2021. Front Immunol. 2021. PMID: 34777377 Free PMC article. Review.
-
ESRP1 drives subtype-specific breast cancer progression through ER-regulated transcriptional programs and EMT-related splicing switch.Am J Cancer Res. 2025 Jun 25;15(6):2807-2825. doi: 10.62347/OXPE5390. eCollection 2025. Am J Cancer Res. 2025. PMID: 40667548 Free PMC article.
-
The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing.Nat Commun. 2020 Jan 24;11(1):486. doi: 10.1038/s41467-020-14304-1. Nat Commun. 2020. PMID: 31980632 Free PMC article.
-
The function of alternative splicing in the proteome: rewiring protein interactomes to put old functions into new contexts.Nat Struct Mol Biol. 2023 Dec;30(12):1844-1856. doi: 10.1038/s41594-023-01155-9. Epub 2023 Nov 30. Nat Struct Mol Biol. 2023. PMID: 38036695 Review.
-
ESRP1-regulated isoform switching of LRRFIP2 determines metastasis of gastric cancer.Nat Commun. 2022 Oct 28;13(1):6274. doi: 10.1038/s41467-022-33786-9. Nat Commun. 2022. PMID: 36307405 Free PMC article.
References
-
- Barac A, Basile J, Vázquez-Prado J, Gao Y, Zheng Y, and Gutkind JS (2004). Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J. Biol. Chem 279, 6182–6189. - PubMed
-
- Blencowe BJ (2017). The relationship between alternative splicing and proteomic complexity. Trends Biochem. Sci 42, 407–408. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

