Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges
- PMID: 30486299
- PMCID: PMC6321045
- DOI: 10.3390/ijms19123755
Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges
Abstract
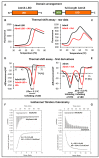
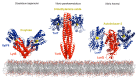
Bacteria possess a large number of signal transduction systems that sense and respond to different environmental cues. Most frequently these are transcriptional regulators, two-component systems and chemosensory pathways. A major bottleneck in the field of signal transduction is the lack of information on signal molecules that modulate the activity of the large majority of these systems. We review here the progress made in the functional annotation of sensor proteins using high-throughput ligand screening approaches of purified sensor proteins or individual ligand binding domains. In these assays, the alteration in protein thermal stability following ligand binding is monitored using Differential Scanning Fluorimetry. We illustrate on several examples how the identification of the sensor protein ligand has facilitated the elucidation of the molecular mechanism of the regulatory process. We will also discuss the use of virtual ligand screening approaches to identify sensor protein ligands. Both approaches have been successfully applied to functionally annotate a significant number of bacterial sensor proteins but can also be used to study proteins from other kingdoms. The major challenge consists in the study of sensor proteins that do not recognize signal molecules directly, but that are activated by signal molecule-loaded binding proteins.
Keywords: bacterial signal transduction systems; chemoreceptors; chemotaxis; sensor kinases; transcriptional regulators.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

