Low Concentration of Antibiotics Modulates Gut Microbiota at Different Levels in Pre-Weaning Dairy Calves
- PMID: 30486334
- PMCID: PMC6313529
- DOI: 10.3390/microorganisms6040118
Low Concentration of Antibiotics Modulates Gut Microbiota at Different Levels in Pre-Weaning Dairy Calves
Abstract
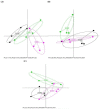
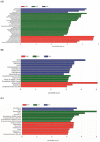
The aim of this study was to investigate the effect of feeding milk replacer (MR) with two different antibiotics treatments on the gut microbiota of pre-weaning calves. Twelve (12) Holstein male calves at 1-day-old were randomly assigned to: milk replacer without antibiotics (CON), milk replacer plus low cocktail of antibiotics (LCA) concentration (penicillin 0.024 mg/L, streptomycin 0.025 mg/L, tetracycline 0.1 mg/L, ceftiofur 0.33 mg/L), and milk replacer plus a low concentration of single antibiotic (LSA; ceftiofur 0.33 mg/L). All the calves were harvested at 35-day-old, and the digesta from the ileum and colon was collected in addition to fecal samples. Samples were analyzed by 16S rRNA gene using Illumina MiSeq platform. Results showed that there were significant differences among treatments in the ileum, where LCA significantly reduced the relative abundance of Enterobacteriaceae (P = 0.02) especially Escherichia-coli (P = 0.02), while LSA significantly reduced the relative abundance of Comamonas (P = 0.02). In the colon and rectum, LSA treatment was significantly enriched with the class Bacilli, whereas the control group was significantly enriched with Alloprevotlla (P = 0.03). However, at the family level in the rectum LCA and LSA significantly reduced the relative abundance of Acidaminococcaceae (P = 0.01). Moreover, at the genera level in the colon, LSA significantly increased Prevotellaceae_Ga6A1_ group (P = 0.02), whereas in the rectum both of treatments reduced the relative abundance of Phascolarctobacterium (P = 0.01). In conclusion, the overall low cocktail of antibiotics concentration induced changes at different taxonomic levels; specifically the decrease in Escherichia-coli which might subsequently reduce the incidences of diarrhea in calves.
Keywords: antibiotics; calf; colon; ileum; microbiota; rectum.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Yeoman C.J., Ishaq S.L., Bichi E., Olivo S.K., Lowe J., Aldridge B.M. Biogeographical Differences in the Influence of Maternal Microbial Sources on the Early Successional Development of the Bovine Neonatal Gastrointestinal tract. Sci. Rep. 2018;8:3197. doi: 10.1038/s41598-018-21440-8. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

