Small Noncoding Regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia Complex
- PMID: 30486355
- PMCID: PMC6321483
- DOI: 10.3390/ijms19123759
Small Noncoding Regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia Complex
Abstract
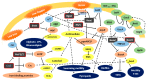
Cystic fibrosis (CF) is the most life-limiting autosomal recessive disorder in Caucasians. CF is characterized by abnormal viscous secretions that impair the function of several tissues, with chronic bacterial airway infections representing the major cause of early decease of these patients. Pseudomonas aeruginosa and bacteria from the Burkholderia cepacia complex (Bcc) are the leading pathogens of CF patients' airways. A wide array of virulence factors is responsible for the success of infections caused by these bacteria, which have tightly regulated responses to the host environment. Small noncoding RNAs (sRNAs) are major regulatory molecules in these bacteria. Several approaches have been developed to study P. aeruginosa sRNAs, many of which were characterized as being involved in the virulence. On the other hand, the knowledge on Bcc sRNAs remains far behind. The purpose of this review is to update the knowledge on characterized sRNAs involved in P. aeruginosa virulence, as well as to compile data so far achieved on sRNAs from the Bcc and their possible roles on bacteria virulence.
Keywords: Burkholderia cepacia complex; Pseudomonas aeruginosa; cystic fibrosis; pathogenicity; small noncoding regulatory RNAs.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article.
-
The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans.Microbiology (Reading). 2010 Mar;156(Pt 3):896-908. doi: 10.1099/mic.0.035139-0. Epub 2009 Nov 26. Microbiology (Reading). 2010. PMID: 19942656
-
Non-coding regulatory sRNAs from bacteria of the Burkholderia cepacia complex.Appl Microbiol Biotechnol. 2024 Apr 2;108(1):280. doi: 10.1007/s00253-024-13121-6. Appl Microbiol Biotechnol. 2024. PMID: 38563885 Free PMC article. Review.
-
The multifarious, multireplicon Burkholderia cepacia complex.Nat Rev Microbiol. 2005 Feb;3(2):144-56. doi: 10.1038/nrmicro1085. Nat Rev Microbiol. 2005. PMID: 15643431 Review.
-
Molecular epidemiology of Pseudomonas aeruginosa, Burkholderia cepacia complex and methicillin-resistant Staphylococcus aureus in a cystic fibrosis center.J Cyst Fibros. 2004 Aug;3(3):159-63. doi: 10.1016/j.jcf.2004.03.010. J Cyst Fibros. 2004. PMID: 15463902
Cited by
-
The Small RNA ErsA Plays a Role in the Regulatory Network of Pseudomonas aeruginosa Pathogenicity in Airway Infections.mSphere. 2020 Oct 14;5(5):e00909-20. doi: 10.1128/mSphere.00909-20. mSphere. 2020. PMID: 33055260 Free PMC article.
-
Conquering the host: Bordetella spp. and Pseudomonas aeruginosa molecular regulators in lung infection.Front Microbiol. 2022 Sep 26;13:983149. doi: 10.3389/fmicb.2022.983149. eCollection 2022. Front Microbiol. 2022. PMID: 36225372 Free PMC article. Review.
-
Specific and Global RNA Regulators in Pseudomonas aeruginosa.Int J Mol Sci. 2021 Aug 11;22(16):8632. doi: 10.3390/ijms22168632. Int J Mol Sci. 2021. PMID: 34445336 Free PMC article. Review.
-
Biofilm Signaling, Composition and Regulation in Burkholderia pseudomallei.J Microbiol Biotechnol. 2023 Jan 28;33(1):15-27. doi: 10.4014/jmb.2207.07032. Epub 2022 Oct 17. J Microbiol Biotechnol. 2023. PMID: 36451302 Free PMC article. Review.
-
The Naphthalene Catabolic Genes of Pseudomonas putida BS3701: Additional Regulatory Control.Front Microbiol. 2020 Jun 5;11:1217. doi: 10.3389/fmicb.2020.01217. eCollection 2020. Front Microbiol. 2020. PMID: 32582120 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

