Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection
- PMID: 30486834
- PMCID: PMC6264784
- DOI: 10.1186/s12967-018-1706-1
Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection
Abstract
Background: The events in early HIV infection (EHI) are important determinants of disease severity and progression rate to AIDS, but the mechanisms of pathogenesis in EHI have not been fully understood. Circular RNAs (circRNAs) have been verified as "microRNA sponges" that regulate gene expression through competing endogenous RNA (ceRNA) networks, but circRNA expression profiles and their contribution to EHI pathogenesis are still unclear.
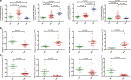
Methods: Two different libraries were constructed with RNA from human peripheral blood mononuclear cells from 3 HARRT-naive EHI patients and 3 healthy controls (HCs). The complete transcriptomes were sequenced with RNA sequencing (RNA-Seq) and miRNA sequencing (miRNA-Seq). The differentially expressed (DE) RNAs were validated with RT-qPCR. The circRNA profile and circRNA-associated-ceRNA network in EHI were analyzed with the integrated data of RNA-Seq and miRNA-Seq. Gene ontology (GO) analysis was used to annotate the circRNAs involved in the circRNA-associated-ceRNA networks.
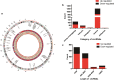
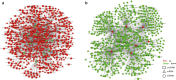
Results: A total of 1365 circRNAs, 30 miRNAs, and 2049 mRNAs were differentially expressed between HARRT-naive EHI patients and HCs. A ceRNA network was constructed with 516 DE circRNAs and 903 DE mRNAs that shared miR response elements with 21 DE miRNAs. GO analysis demonstrated the multiple roles of the circRNAs enriched in EHI with circRNA-associated-ceRNA networks, such as immune response, inflammatory response and defense responses to virus, 67 circRNAs were revealed to be potentially involved in HIV-1 replication through regulating the expression of CCNK, CDKN1A and IL-15.
Conclusions: This study, for the first time, revealed a large circRNA profile and complex pathogenesis roles of circRNAs in EHI. A group of enriched circRNAs and associated circRNA-associated-ceRNA networks might contribute to HIV replication regulation and provide novel potential targets for both the pathogenesis of EHI and antiviral therapy.
Keywords: HIV-1; Viral replication; ceRNA; circRNA.
Figures

Similar articles
-
Integrated analysis of lncRNA, miRNA and mRNA profiles reveals potential lncRNA functions during early HIV infection.J Transl Med. 2021 Apr 1;19(1):135. doi: 10.1186/s12967-021-02802-9. J Transl Med. 2021. PMID: 33794921 Free PMC article.
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Identification and characterization of circular RNAs in the silkworm midgut following Bombyx mori cytoplasmic polyhedrosis virus infection.RNA Biol. 2018 Feb 1;15(2):292-301. doi: 10.1080/15476286.2017.1411461. Epub 2017 Dec 21. RNA Biol. 2018. PMID: 29268657 Free PMC article.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
-
The circRNA-miRNA-mRNA regulatory network in plasma and peripheral blood mononuclear cells and the potential associations with the pathogenesis of systemic lupus erythematosus.Clin Rheumatol. 2023 Jul;42(7):1885-1896. doi: 10.1007/s10067-023-06560-5. Epub 2023 Mar 2. Clin Rheumatol. 2023. PMID: 36862342 Review.
Cited by
-
Hsa_circRNA_0001971 contributes to oral squamous cell carcinoma progression via miR-186-5p/Fibronectin type III domain containing 3B axis.J Clin Lab Anal. 2022 Mar;36(3):e24245. doi: 10.1002/jcla.24245. Epub 2022 Jan 21. J Clin Lab Anal. 2022. PMID: 35060189 Free PMC article.
-
Genome-wide analysis of differentially expressed profiles of mRNAs, lncRNAs and circRNAs in chickens during Eimeria necatrix infection.Parasit Vectors. 2020 Apr 3;13(1):167. doi: 10.1186/s13071-020-04047-9. Parasit Vectors. 2020. PMID: 32245514 Free PMC article.
-
Gene dysregulation in acute HIV-1 infection - early transcriptomic analysis reveals the crucial biological functions affected.Front Cell Infect Microbiol. 2023 Apr 3;13:1074847. doi: 10.3389/fcimb.2023.1074847. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37077524 Free PMC article.
-
Comprehensive circRNA-microRNA-mRNA network analysis revealed the novel regulatory mechanism of Trichosporon asahii infection.Mil Med Res. 2021 Mar 9;8(1):19. doi: 10.1186/s40779-021-00311-w. Mil Med Res. 2021. PMID: 33750466 Free PMC article.
-
Emerging Roles of Extracellular Vesicle-Delivered Circular RNAs in Atherosclerosis.Front Cell Dev Biol. 2022 Apr 4;10:804247. doi: 10.3389/fcell.2022.804247. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35445015 Free PMC article. Review.
References
-
- Powers KA, Ghani AC, Miller WC, Hoffman IF, Pettifor AE, Kamanga G, et al. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet. 2011;378(9787):256–268. doi: 10.1016/S0140-6736(11)60842-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2017ZX10201101/the Mega-Projects of National Science Research for the 13th Five-Year Plan/International
- 81371787/National Natural Science Foundation of China/International
- IRT_16R70/Innovation Team Development Program of the Ministry of Education/International
- 2018PT31042/Central Public-interest Scientific Institution Basal Research Fund/International
LinkOut - more resources
Full Text Sources
Medical

