Molecular identification of Plasmodium species responsible for malaria reveals Plasmodium vivax isolates in Duffy negative individuals from southwestern Nigeria
- PMID: 30486887
- PMCID: PMC6263541
- DOI: 10.1186/s12936-018-2588-7
Molecular identification of Plasmodium species responsible for malaria reveals Plasmodium vivax isolates in Duffy negative individuals from southwestern Nigeria
Abstract
Background: Malaria in Nigeria is principally due to Plasmodium falciparum and, to a lesser extent to Plasmodium malariae and Plasmodium ovale. Plasmodium vivax is thought to be absent in Nigeria in particular and sub-Saharan Africa in general, due to the near fixation of the Duffy negative gene in this population. Nevertheless, there are frequent reports of P. vivax infection in Duffy negative individuals in the sub-region, including reports from two countries sharing border with Nigeria to the west (Republic of Benin) and east (Cameroon). Additionally, there were two cases of microscopic vivax-like malaria from Nigerian indigenous population. Hence molecular surveillance of the circulating Plasmodium species in two states (Lagos and Edo) of southwestern Nigeria was carried out.
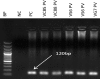
Methods: A cross-sectional survey between September 2016 and March 2017 was conducted. 436 febrile patients were included for the present work. Venous blood of these patients was subjected to RDT as well as microscopy. Further, parasite DNA was isolated from positive samples and PCR diagnostic was employed followed by direct sequencing of the 18S rRNA of Plasmodium species as well as sequencing of a portion of the promoter region of the Duffy antigen receptor for chemokines. Samples positive for P. vivax were re-amplified several times and finally using the High Fidelity Taq to rule out any bias introduced.
Results: Of the 256 (58.7%) amplifiable malaria parasite DNA, P. falciparum was, as expected, the major cause of infection, either alone 85.5% (219/256; 97 from Edo and 122 from Lagos), or mixed with P. malariae 6.3% (16/256) or with P. vivax 1.6% (4/256). Only one of the five P. vivax isolates was found to be a single infection. DNA sequencing and subsequent alignment of the 18S rRNA of P. vivax with the reference strains displayed very high similarities (100%). Remarkably, the T-33C was identified in all P. vivax samples, thus confirming that all vivax-infected patients in the current study are Duffy negative.
Conclusion: The present study gave the first molecular evidence of P. vivax in Nigeria in Duffy negative individuals. Though restricted to two states; Edo in South-South and Lagos in South-west Nigeria, the real burden of this species in Nigeria and sub-Saharan Africa might have been underestimated, hence there is need to put in place a country-wide, as well as a sub-Saharan Africa-wide surveillance and appropriate control measures.
Keywords: Duffy negative; Edo; Lagos; Malaria; Plasmodium vivax; Sub-Saharan Africa.
Figures



References
-
- WHO. World Malaria Report 2016. Geneva: World Health Organization; 2016. http://www.who.int/malaria/publications/world_malaria_report_2013/en/. Accessed 3 Sept 2017.
-
- WHO. World Malaria Report 2017. Geneva: World Health Organization; 2017. 10.1071/EC12504. Accessed 3 Mar 2018.
-
- Federal Ministry of Health. National guideline for diagnosis and treatment of malaria, 3rd edn; 2015. Accessed 22 Dec 2016.
-
- Mahende C, Ngasala B, Lusingu J, Yong TS, Lushino P, Lemnge M, et al. Performance of rapid diagnostic test, blood-film microscopy and PCR for the diagnosis of malaria infection among febrile children from Korogwe District, Tanzania. Malar J. 2016;15:391. doi: 10.1186/s12936-016-1450-z. - DOI - PMC - PubMed
-
- Abeku TA, Kristan M, Jones C, Beard J, Mueller DH, Okia M, et al. Determinants of the accuracy of rapid diagnostic tests in malaria case management: evidence from low and moderate transmission settings in the East African highlands. Malar J. 2008;7:202. doi: 10.1186/1475-2875-7-202. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

