SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines
- PMID: 30487218
- PMCID: PMC6294928
- DOI: 10.1073/pnas.1716349115
SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines
Abstract
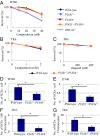
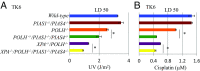
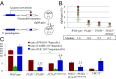
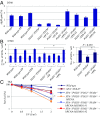
DNA damage tolerance (DDT) releases replication blockage caused by damaged nucleotides on template strands employing two alternative pathways, error-prone translesion DNA synthesis (TLS) and error-free template switch (TS). Lys164 of proliferating cell nuclear antigen (PCNA) is SUMOylated during the physiological cell cycle. To explore the role for SUMOylation of PCNA in DDT, we characterized chicken DT40 and human TK6 B cells deficient in the PIAS1 and PIAS4 small ubiquitin-like modifier (SUMO) E3 ligases. DT40 cells have a unique advantage in the phenotypic analysis of DDT as they continuously diversify their immunoglobulin (Ig) variable genes by TLS and TS [Ig gene conversion (GC)], both relieving replication blocks at abasic sites without accompanying by DNA breakage. Remarkably, PIAS1-/-/PIAS4-/- cells displayed a multifold decrease in SUMOylation of PCNA at Lys164 and over a 90% decrease in the rate of TS. Likewise, PIAS1-/-/PIAS4-/- TK6 cells showed a shift of DDT from TS to TLS at a chemosynthetic UV lesion inserted into the genomic DNA. The PCNAK164R/K164R mutation caused a ∼90% decrease in the rate of Ig GC and no additional impact on PIAS1-/-/PIAS4-/- cells. This epistatic relationship between the PCNAK164R/K164R and the PIAS1-/-/PIAS4-/- mutations suggests that PIAS1 and PIAS4 promote TS mainly through SUMOylation of PCNA at Lys164. This idea is further supported by the data that overexpression of a PCNA-SUMO1 chimeric protein restores defects in TS in PIAS1-/-/PIAS4-/- cells. In conclusion, SUMOylation of PCNA at Lys164 promoted by PIAS1 and PIAS4 ensures the error-free release of replication blockage during physiological DNA replication in metazoan cells.
Keywords: PCNA; PIAS1; PIAS4; SUMOylation; template switching (TS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
PDIP38/PolDIP2 controls the DNA damage tolerance pathways by increasing the relative usage of translesion DNA synthesis over template switching.PLoS One. 2019 Mar 6;14(3):e0213383. doi: 10.1371/journal.pone.0213383. eCollection 2019. PLoS One. 2019. PMID: 30840704 Free PMC article.
-
Multisite SUMOylation restrains DNA polymerase η interactions with DNA damage sites.J Biol Chem. 2020 Jun 19;295(25):8350-8362. doi: 10.1074/jbc.RA120.013780. Epub 2020 Apr 29. J Biol Chem. 2020. PMID: 32350109 Free PMC article.
-
A genetic study based on PCNA-ubiquitin fusions reveals no requirement for PCNA polyubiquitylation in DNA damage tolerance.DNA Repair (Amst). 2017 Jun;54:46-54. doi: 10.1016/j.dnarep.2017.04.003. Epub 2017 Apr 14. DNA Repair (Amst). 2017. PMID: 28458162
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
Cited by
-
Division of labor of Y-family polymerases in translesion-DNA synthesis for distinct types of DNA damage.PLoS One. 2021 Jun 1;16(6):e0252587. doi: 10.1371/journal.pone.0252587. eCollection 2021. PLoS One. 2021. PMID: 34061890 Free PMC article.
-
Enhancing the sensitivity of the thymidine kinase assay by using DNA repair-deficient human TK6 cells.Environ Mol Mutagen. 2020 Jul;61(6):602-610. doi: 10.1002/em.22371. Epub 2020 Apr 15. Environ Mol Mutagen. 2020. PMID: 32243652 Free PMC article.
-
Mechanisms of damage tolerance and repair during DNA replication.Nucleic Acids Res. 2021 Apr 6;49(6):3033-3047. doi: 10.1093/nar/gkab101. Nucleic Acids Res. 2021. PMID: 33693881 Free PMC article. Review.
-
The Dark Side of UV-Induced DNA Lesion Repair.Genes (Basel). 2020 Dec 2;11(12):1450. doi: 10.3390/genes11121450. Genes (Basel). 2020. PMID: 33276692 Free PMC article. Review.
-
Mechanisms of direct replication restart at stressed replisomes.DNA Repair (Amst). 2020 Nov;95:102947. doi: 10.1016/j.dnarep.2020.102947. Epub 2020 Aug 16. DNA Repair (Amst). 2020. PMID: 32853827 Free PMC article. Review. No abstract available.
References
-
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. - PubMed
-
- Pommier Y, O’Connor MJ, de Bono J, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

