Preclinical development of a microRNA-based therapy for intervertebral disc degeneration
- PMID: 30487517
- PMCID: PMC6262020
- DOI: 10.1038/s41467-018-07360-1
Preclinical development of a microRNA-based therapy for intervertebral disc degeneration
Abstract
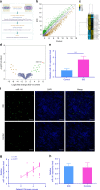
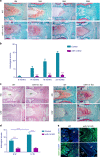
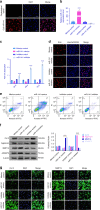
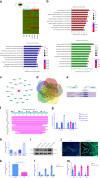
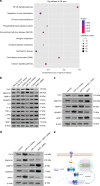
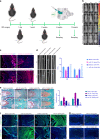
Understanding the molecular mechanisms regulating the maintenance and destruction of intervertebral disc may lead to the development of new therapies for intervertebral disc degeneration (IDD). Here we present evidence from miRNA microarray analyses of clinical data sets along with in vitro and in vivo experiments that miR-141 is a key regulator of IDD. Gain- and loss-of-function studies show that miR-141 drives IDD by inducing nucleus pulposus (NP) apoptosis. Furthermore, miR-141 KO in mice attenuated spontaneous and surgically induced IDD. Mechanistically, miR-141 promotes IDD development by targeting and depleting SIRT1, a negative regulator of NF-κB pathway. Therapeutically, upregulation or downregulation of miR-141 by nanoparticle delivery in IDD model aggravated or alleviated experimental IDD, respectively. Our findings reveal a novel mechanism by which miR-141, in part, promotes IDD progression by interacting with SIRT1/NF-κB pathway. Blockade of miR-141 in vivo may serve as a potential therapeutic approach in the treatment of IDD.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Platelet-rich plasma-derived extracellular vesicles inhibit NF-κB/NLRP3 pathway-mediated pyroptosis in intervertebral disc degeneration via the MALAT1/microRNA-217/SIRT1 axis.Cell Signal. 2024 May;117:111106. doi: 10.1016/j.cellsig.2024.111106. Epub 2024 Feb 17. Cell Signal. 2024. PMID: 38373669
-
CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis.EBioMedicine. 2020 Mar;53:102679. doi: 10.1016/j.ebiom.2020.102679. Epub 2020 Feb 26. EBioMedicine. 2020. PMID: 32114390 Free PMC article.
-
Effective delivery of miR-150-5p with nucleus pulposus cell-specific nanoparticles attenuates intervertebral disc degeneration.J Nanobiotechnology. 2024 May 27;22(1):292. doi: 10.1186/s12951-024-02561-x. J Nanobiotechnology. 2024. PMID: 38802882 Free PMC article.
-
MicroRNA in intervertebral disc degeneration.Cell Prolif. 2015 Jun;48(3):278-83. doi: 10.1111/cpr.12180. Epub 2015 Mar 4. Cell Prolif. 2015. PMID: 25736871 Free PMC article. Review.
-
A Bioinformatic Analysis of MicroRNAs' Role in Human Intervertebral Disc Degeneration.Pain Med. 2019 Dec 1;20(12):2459-2471. doi: 10.1093/pm/pnz015. Pain Med. 2019. PMID: 30953590 Review.
Cited by
-
Dysregulation of MicroRNAs in Hypertrophy and Ossification of Ligamentum Flavum: New Advances, Challenges, and Potential Directions.Front Genet. 2021 Apr 12;12:641575. doi: 10.3389/fgene.2021.641575. eCollection 2021. Front Genet. 2021. PMID: 33912216 Free PMC article.
-
Exosomes from M2c macrophages alleviate intervertebral disc degeneration by promoting synthesis of the extracellular matrix via MiR-124/CILP/TGF-β.Bioeng Transl Med. 2023 Feb 16;8(6):e10500. doi: 10.1002/btm2.10500. eCollection 2023 Nov. Bioeng Transl Med. 2023. PMID: 38023721 Free PMC article.
-
The potential mechanisms and application prospects of non-coding RNAs in intervertebral disc degeneration.Front Endocrinol (Lausanne). 2022 Dec 8;13:1081185. doi: 10.3389/fendo.2022.1081185. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36568075 Free PMC article. Review.
-
Regulatory roles of noncoding RNAs in intervertebral disc degeneration as potential therapeutic targets (Review).Exp Ther Med. 2022 Dec 1;25(1):44. doi: 10.3892/etm.2022.11743. eCollection 2023 Jan. Exp Ther Med. 2022. PMID: 36569433 Free PMC article. Review.
-
Polydopamine Nanoparticles Targeting Ferroptosis Mitigate Intervertebral Disc Degeneration Via Reactive Oxygen Species Depletion, Iron Ions Chelation, and GPX4 Ubiquitination Suppression.Adv Sci (Weinh). 2023 May;10(13):e2207216. doi: 10.1002/advs.202207216. Epub 2023 Mar 23. Adv Sci (Weinh). 2023. PMID: 36951540 Free PMC article.
References
-
- Abajobir AA, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. - DOI - PMC - PubMed
-
- Vos T, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. - DOI - PMC - PubMed
-
- Roberts S, et al. Histology and pathology of the human intervertebral disc. J. Bone Joint Surg. Am. 2006;88(S2):10–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

