Selective expansion of myeloid and NK cells in humanized mice yields human-like vaccine responses
- PMID: 30487575
- PMCID: PMC6262001
- DOI: 10.1038/s41467-018-07478-2
Selective expansion of myeloid and NK cells in humanized mice yields human-like vaccine responses
Abstract
Mice engrafted with components of a human immune system have become widely-used models for studying aspects of human immunity and disease. However, a defined methodology to objectively measure and compare the quality of the human immune response in different models is lacking. Here, by taking advantage of the highly immunogenic live-attenuated yellow fever virus vaccine YFV-17D, we provide an in-depth comparison of immune responses in human vaccinees, conventional humanized mice, and second generation humanized mice. We demonstrate that selective expansion of human myeloid and natural killer cells promotes transcriptomic responses akin to those of human vaccinees. These enhanced transcriptomic profiles correlate with the development of an antigen-specific cellular and humoral response to YFV-17D. Altogether, our approach provides a robust scoring of the quality of the human immune response in humanized mice and highlights a rational path towards developing better pre-clinical models for studying the human immune response and disease.
Conflict of interest statement
The authors declare no competing interests.
Figures


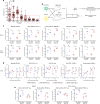
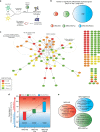




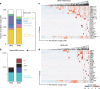
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL095791/HL/NHLBI NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 AI079031/AI/NIAID NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- R33 CA202820/CA/NCI NIH HHS/United States
- T32 GM007388/GM/NIGMS NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- U24 AI118672/AI/NIAID NIH HHS/United States
- U54 CA217377/CA/NCI NIH HHS/United States
- R01 AI138797/AI/NIAID NIH HHS/United States
- R01 HL134539/HL/NHLBI NIH HHS/United States
- RM1 HG006193/HG/NHGRI NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

