PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation
- PMID: 30487600
- PMCID: PMC9402428
- DOI: 10.1038/s41586-018-0761-3
PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation
Abstract
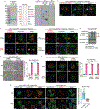
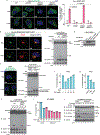
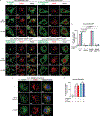
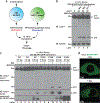
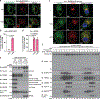
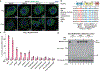
The NLRP3 inflammasome, which has been linked to human inflammatory diseases, is activated by diverse stimuli. How these stimuli activate NLRP3 is unknown. Here we show that different NLRP3 stimuli lead to disassembly of the trans-Golgi network (TGN). NLRP3 is recruited to the dispersed TGN (dTGN) through ionic bonding between its conserved polybasic region and negatively charged phosphatidylinositol-4-phosphate (PtdIns4P) on the dTGN. The dTGN then serves as a scaffold for NLRP3 aggregation into multiple puncta, leading to polymerization of the adaptor protein ASC, thereby activating the downstream signalling cascade. Disruption of the interaction between NLRP3 and PtdIns4P on the dTGN blocked NLRP3 aggregation and downstream signalling. These results indicate that recruitment of NLRP3 to dTGN is an early and common cellular event that leads to NLRP3 aggregation and activation in response to diverse stimuli.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
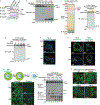

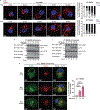
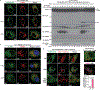

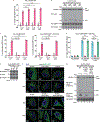
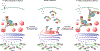

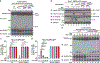
Comment in
-
Trans-Golgi network breaks away to activate NLRP3.Nat Rev Immunol. 2019 Feb;19(2):68-69. doi: 10.1038/s41577-018-0111-6. Nat Rev Immunol. 2019. PMID: 30602732 No abstract available.
-
Divide to conquer: NLRP3 is activated on dispersed trans-Golgi network.Cell Res. 2019 Mar;29(3):181-182. doi: 10.1038/s41422-018-0138-z. Cell Res. 2019. PMID: 30664729 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

