Two Novel KRIT1 and CCM2 Mutations in Patients Affected by Cerebral Cavernous Malformations: New Information on CCM2 Penetrance
- PMID: 30487773
- PMCID: PMC6246743
- DOI: 10.3389/fneur.2018.00953
Two Novel KRIT1 and CCM2 Mutations in Patients Affected by Cerebral Cavernous Malformations: New Information on CCM2 Penetrance
Abstract
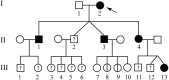
Wide comprehension of genetic features of cerebral cavernous malformations (CCM) represents the starting point to better manage patients and risk rating in relatives. The causative mutations spectrum is constantly growing. KRIT1, CCM2, and PDCD10 are the three loci to date linked to familial CCM development, although germline mutations have also been detected in patients affected by sporadic forms. In this context, the main challenge is to draw up criteria to formulate genotype-phenotype correlations. Clearly, genetic factors determining incomplete penetrance of CCM need to be identified. Here, we report two novel intronic variants probably affecting splicing. Molecular screening of CCM genes was performed on DNA purified by peripheral blood. Coding exons and intron-exon boundaries were sequenced by the Sanger method. The first was detected in a sporadic patient and involves KRIT1. The second affects CCM2 and it is harbored by a woman with familial CCM. Interestingly, molecular analysis extended to both healthy and ill relatives allowed to estimate, for the first time, a penetrance for CCM2 lower than 100%, as to date reported. Moreover, heterogeneity of clinical manifestations among those affected carrying the same genotype further confirms involvement of modifier factors in CCM development.
Keywords: CCM; CCM genotype-phenotype correlation; CCM2 penetrance; pathogenesis; splicing variants.
Figures



References
LinkOut - more resources
Full Text Sources

