In silico Design of Laccase Thermostable Mutants From Lacc 6 of Pleurotus Ostreatus
- PMID: 30487785
- PMCID: PMC6247816
- DOI: 10.3389/fmicb.2018.02743
In silico Design of Laccase Thermostable Mutants From Lacc 6 of Pleurotus Ostreatus
Abstract
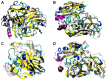
Fungal laccase enzymes have a great biotechnological potential for bioremediation processes due to their ability to degrade compounds such as ρ-diphenol, aminophenols, polyphenols, polyamines, and aryldiamines. These enzymes have activity at different pH and temperature values, however, high temperatures can cause partial or total loss of enzymatic activity, so it is appropriate to do research to modify their secondary and/or tertiary structure to make them more resistant to extreme temperature conditions. In silico, a structure of the Lacc 6 enzyme of Pleurotus ostreatus was constructed using a laccase of Trametes versicolor as a template. From this structure, 16 mutants with possible resistance at high temperature due to ionic interactions, salt bridges and disulfide bonds were also obtained in silico. It was determined that 12 mutants called 4-DB, 3-DB, D233C-T310C, F468P, 3-SB, L132T, N79D, N372D, P203C, P203V, T147E, and W85F, presented the lowest thermodynamic energy. Based on the previous criterion and determining the least flexibility in the protein structures, three mutants (4-DB, 3-DB, and P203C) were selected, which may present high stability at high temperatures without affecting their active site. The obtained results allow the understanding of the molecular base that increase the structural stability of the enzyme Lacc 6 of Pleurotus ostreatus, achieving the in silico generation of mutants, which could have activity at high temperatures.
Keywords: Lacc 6; Pleurotus ostreatus; energy minimization; laccase; mutants.
Figures






Similar articles
-
Effect of Pleurotus ostreatus and Trametes versicolor on triclosan biodegradation and activity of laccase and manganese peroxidase enzymes.Microb Pathog. 2020 Dec;149:104473. doi: 10.1016/j.micpath.2020.104473. Epub 2020 Sep 8. Microb Pathog. 2020. PMID: 32916239
-
Temperature affects the production, activity and stability of ligninolytic enzymes in Pleurotus ostreatus and Trametes versicolor.Folia Microbiol (Praha). 2007;52(5):498-502. doi: 10.1007/BF02932110. Folia Microbiol (Praha). 2007. PMID: 18298047
-
Estimation of bound and free fractions of lignocellulose-degrading enzymes of wood-rotting fungi Pleurotus ostreatus, Trametes versicolor and Piptoporus betulinus.Res Microbiol. 2006 Mar;157(2):119-24. doi: 10.1016/j.resmic.2005.06.004. Epub 2005 Aug 8. Res Microbiol. 2006. PMID: 16125911
-
Random mutants of a Pleurotus ostreatus laccase as new biocatalysts for industrial effluents bioremediation.J Appl Microbiol. 2010 Mar;108(3):998-1006. doi: 10.1111/j.1365-2672.2009.04505.x. Epub 2009 Jul 30. J Appl Microbiol. 2010. PMID: 19735323
-
Essential role of the N- and C-terminals of laccase from Pleurotus florida on the laccase activity and stability.Appl Biochem Biotechnol. 2014 Nov;174(5):2007-17. doi: 10.1007/s12010-014-1147-0. Epub 2014 Aug 27. Appl Biochem Biotechnol. 2014. PMID: 25161036
Cited by
-
Altering the Properties of Laccases from Ensifer meliloti (Sinorhizobium meliloti) and Cerrena unicolor by Chemical Modifications of Proteins.Biomolecules. 2025 Apr 4;15(4):531. doi: 10.3390/biom15040531. Biomolecules. 2025. PMID: 40305261 Free PMC article.
-
Laccases: structure, function, and potential application in water bioremediation.Microb Cell Fact. 2019 Nov 14;18(1):200. doi: 10.1186/s12934-019-1248-0. Microb Cell Fact. 2019. PMID: 31727078 Free PMC article. Review.
References
-
- Berka R. M., Schneider P., Golightly E. J., Brown S. H., Madden M., Brown K. M., et al. (1997). Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 63 3151–3157. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

