Site-Specific N-Glycan Characterization of Grass Carp Serum IgM
- PMID: 30487799
- PMCID: PMC6246689
- DOI: 10.3389/fimmu.2018.02645
Site-Specific N-Glycan Characterization of Grass Carp Serum IgM
Abstract
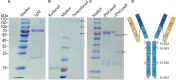
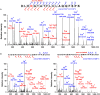
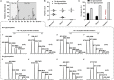
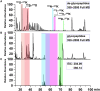
Immunoglobulin M (IgM) is the major antibody in teleost fish and plays an important role in humoral adaptive immunity. The N-linked carbohydrates presenting on IgM have been well documented in higher vertebrates, but little is known regarding site-specific N-glycan characteristics in teleost IgM. In order to characterize these site-specific N-glycans, we conducted the first study of the N-glycans of each glycosylation site of the grass carp serum IgM. Among the four glycosylation sites, the Asn-262, Asn-303, and Asn-426 residues were efficiently glycosylated, while Asn-565 at the C-terminal tailpiece was incompletely occupied. A striking decrease in the level of occupancy at the Asn-565 glycosite was observed in dimeric IgM compared to that in monomeric IgM, and no glycan occupancy of Asn-565 was observed in tetrameric IgM. Glycopeptide analysis with liquid chromatography-electrospray ionization tandem mass spectrometry revealed mainly complex-type glycans with substantial heterogeneity, with neutral; monosialyl-, disialyl- and trisialylated; and fucosyl-and non-fucosyl-oligosaccharides conjugated to grass carp serum IgM. Glycan variation at a single site was greatest at the Asn-262 glycosite. Unlike IgMs in other species, only traces of complex-type and no high-mannose glycans were found at the Asn-565 glycosite. Matrix-assisted laser desorption ionization analysis of released glycans confirmed the overwhelming majority of carbohydrates were of the complex-type. These results indicate that grass carp serum IgM exhibits unique N-glycan features and highly processed oligosaccharides attached to individual glycosites.
Keywords: N-glycan; glycosylation; grass carp; immunoglobulin M; liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS); matrix assisted laser desorption/ionization-time-of-flight-MS (MALDI-TOF-MS); teleost.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

