The Host Microbiota Contributes to Early Protection Against Lung Colonization by Mycobacterium tuberculosis
- PMID: 30487801
- PMCID: PMC6246741
- DOI: 10.3389/fimmu.2018.02656
The Host Microbiota Contributes to Early Protection Against Lung Colonization by Mycobacterium tuberculosis
Abstract
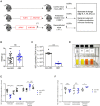
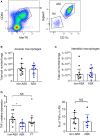
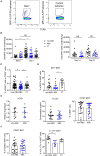
Tuberculosis (TB), caused by the airborne bacterial pathogen Mycobacterium tuberculosis, remains a major source of morbidity and mortality worldwide. So far, the study of host-pathogen interactions in TB has mostly focused on the physiology and virulence of the pathogen, as well as, on the various innate and adaptive immune compartments of the host. Microbial organisms endogenous to our body, the so-called microbiota, interact not only with invading pathogens, but also with our immune system. Yet, the impact of the microbiota on host defense against M. tuberculosis remains poorly understood. In order to address this question, we adapted a robust and reproducible mouse model of microbial dysbiosis based on a combination of wide-spectrum antibiotics. We found that microbiota dysbiosis resulted in an increased early colonization of the lungs by M. tuberculosis during the first week of infection, correlating with an altered diversity of the gut microbiota during this time period. At the cellular level, no significant difference in the recruitment of conventional myeloid cells, including macrophages, dendritic cells and neutrophils, to the lungs could be detected during the first week of infection between microbiota-competent and -deficient mice. At the molecular level, microbiota depletion did not impact the global production of pro-inflammatory cytokines, such as interferon (IFN)γ, tumor necrosis factor (TNF)α and interleukin (IL)-1β in the lungs. Strikingly, a reduced number of mucosal-associated invariant T (MAIT) cells, a population of innate-like lymphocytes whose development is known to depend on the host microbiota, was observed in the lungs of the antibiotics-treated animals after 1week of infection. These cells produced less IL-17A in antibiotics-treated mice. Notably, dysbiosis correction through the inoculation of a complex microbiota in antibiotics-treated animals reversed these phenotypes and improved the ability of MAIT cells to proliferate. Altogether, our results demonstrate that the host microbiota contributes to early protection of lung colonization by M. tuberculosis, possibly through sustaining the function(s) of MAIT cells. Our study calls for a better understanding of the impact of the microbiota on host-pathogen interactions in TB. Ultimately, this study may help to develop novel therapeutic approaches based on the use of beneficial microbes, or components thereof, to boost anti-mycobacterial immunity.
Keywords: IL-17; MAIT cells; macrophage; microbiota; tuberculosis.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

