Modeling Meets Metabolomics-The WormJam Consensus Model as Basis for Metabolic Studies in the Model Organism Caenorhabditis elegans
- PMID: 30488036
- PMCID: PMC6246695
- DOI: 10.3389/fmolb.2018.00096
Modeling Meets Metabolomics-The WormJam Consensus Model as Basis for Metabolic Studies in the Model Organism Caenorhabditis elegans
Abstract
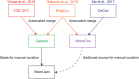
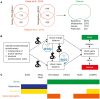
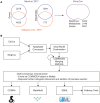
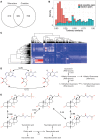
Metabolism is one of the attributes of life and supplies energy and building blocks to organisms. Therefore, understanding metabolism is crucial for the understanding of complex biological phenomena. Despite having been in the focus of research for centuries, our picture of metabolism is still incomplete. Metabolomics, the systematic analysis of all small molecules in a biological system, aims to close this gap. In order to facilitate such investigations a blueprint of the metabolic network is required. Recently, several metabolic network reconstructions for the model organism Caenorhabditis elegans have been published, each having unique features. We have established the WormJam Community to merge and reconcile these (and other unpublished models) into a single consensus metabolic reconstruction. In a series of workshops and annotation seminars this model was refined with manual correction of incorrect assignments, metabolite structure and identifier curation as well as addition of new pathways. The WormJam consensus metabolic reconstruction represents a rich data source not only for in silico network-based approaches like flux balance analysis, but also for metabolomics, as it includes a database of metabolites present in C. elegans, which can be used for annotation. Here we present the process of model merging, correction and curation and give a detailed overview of the model. In the future it is intended to expand the model toward different tissues and put special emphasizes on lipid metabolism and secondary metabolism including ascaroside metabolism in accordance to their central role in C. elegans physiology.
Keywords: Caenorhabditis elegans; metabolic pathways; metabolic reconstruction; metabolism; metabolomics.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

