Lymphotoxin-β receptor-NIK signaling induces alternative RELB/NF-κB2 activation to promote metastatic gene expression and cell migration in head and neck cancer
- PMID: 30488488
- PMCID: PMC7066987
- DOI: 10.1002/mc.22938
Lymphotoxin-β receptor-NIK signaling induces alternative RELB/NF-κB2 activation to promote metastatic gene expression and cell migration in head and neck cancer
Abstract
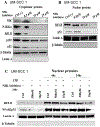
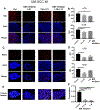
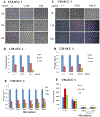
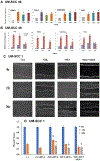
Head and neck squamous cell carcinomas (HNSCC) preferentially spread to regional cervical tissues and lymph nodes. Here, we hypothesized that lymphotoxin-β (LTβ), receptor LTβR, and NF-κB-inducing kinase (NIK), promote the aberrant activation of alternative NF-κB2/RELB pathway and genes, that enhance migration and invasion of HNSCC. Genomic and expression alterations of the alternative NF-kB pathway were examined in 279 HNSCC tumors from The Cancer Genome Atlas (TCGA) and a panel of HNSCC lines. LTβR is amplified or overexpressed in HNSCC of the larynx or oral cavity, while LTβ, NIK, and RELB are overexpressed in cancers arising within lymphoid oropharyngeal and tonsillar sites. Similarly, subsets of HNSCC lines displayed overexpression of LTβR, NIK, and RELB proteins. Recombinant LTβ, and siRNA depletion of endogenous LTβR and NIK, modulated expression of LTβR, NIK, and nuclear translocation of NF-κB2(p52)/RELB as well as functional NF-κB promoter reporter activity. Treatment with a NIK inhibitor (1,3[2H,4H]-Iso-Quinoline Dione) reduced the protein expression of NIK and NF-κB2(p52)/RELB, and blocked LTβ induced nuclear translocation of RELB. NIK and RELB siRNA knockdown or NIK inhibitor slowed HNSCC migration or invation in vitro. LTβ-induces expression of migration and metastasis related genes, including hepatocyte growth/scatter factor receptor MET. Knockdown of NIK or MET similarly inhibited the migration of HNSCC cell lines. This may help explain why HNSCC preferentially migrate to local lymph nodes, where LTβ is expressed. Our findings show that LTβ/LTβR promotes activation of the alternative NIK-NF-κB2/RELB pathway to enhance MET-mediated cell migration in HNSCC, which could be potential therapeutic targets in HNSCC.
Keywords: HNSCC; LTβ/LTβ receptor; NIK; RELB/NF-κB2; migration.
© 2018 Wiley Periodicals, Inc.
Conflict of interest statement
Figures


References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016. Jan-Feb;66(1):7–30. PubMed PMID: 26742998. - PubMed
-
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011. January;11(1):9–22. PubMed PMID: 21160525. - PubMed
-
- Shang Xie HX, Shan Xiaofeng, Liu Baozhong, Wang Kan, Cai* Zhigang. Clinicopathological and prognostic significance of survivin expression in patients with oral squamous cell carcinoma: evidence from a meta-analysis. PLOS One. 2015; 10(2):0116517. PubMed PMID: 25710884. Pubmed Central PMCID: PMC4339736. - PMC - PubMed
-
- Bauer J NS, Reisinger F, Zoller J, Yuan D,Heikenwalder M. Lymphotoxin, NF-kB, and cancer: the dark side of cytokines. Dig Dis. 2012;30(5):453–68. PubMed PMID: 23108301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

