Evolutionary pathways of repeat protein topology in bacterial outer membrane proteins
- PMID: 30489257
- PMCID: PMC6340704
- DOI: 10.7554/eLife.40308
Evolutionary pathways of repeat protein topology in bacterial outer membrane proteins
Abstract
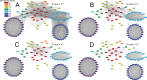
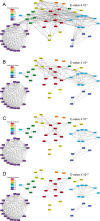
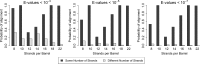
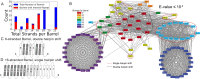
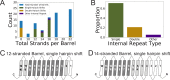
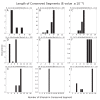
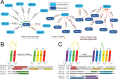
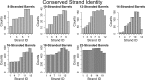
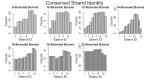
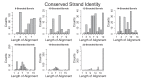
Outer membrane proteins (OMPs) are the proteins in the surface of Gram-negative bacteria. These proteins have diverse functions but a single topology: the β-barrel. Sequence analysis has suggested that this common fold is a β-hairpin repeat protein, and that amplification of the β-hairpin has resulted in 8-26-stranded barrels. Using an integrated approach that combines sequence and structural analyses, we find events in which non-amplification diversification also increases barrel strand number. Our network-based analysis reveals strand-number-based evolutionary pathways, including one that progresses from a primordial 8-stranded barrel to 16-strands and further, to 18-strands. Among these pathways are mechanisms of strand number accretion without domain duplication, like a loop-to-hairpin transition. These mechanisms illustrate perpetuation of repeat protein topology without genetic duplication, likely induced by the hydrophobic membrane. Finally, we find that the evolutionary trace is particularly prominent in the C-terminal half of OMPs, implicating this region in the nucleation of OMP folding.
Keywords: E. coli; beta barrels; beta hairpin; evolutionary biology; molecular biophysics; outer membrane beta barrels; outer membrane proteins; repeat proteins; structural biology.
© 2018, Franklin et al.
Conflict of interest statement
MF, SN, RF, RK, JS No competing interests declared, NB Reviewing editor, eLife
Figures




Comment in
- doi: 10.7554/eLife.44076
Similar articles
-
Evolutionary Engineering a Larger Porin Using a Loop-to-Hairpin Mechanism.J Mol Biol. 2023 Nov 15;435(22):168292. doi: 10.1016/j.jmb.2023.168292. Epub 2023 Sep 26. J Mol Biol. 2023. PMID: 37769963 Free PMC article.
-
Evolution of outer membrane beta-barrels from an ancestral beta beta hairpin.Mol Biol Evol. 2010 Jun;27(6):1348-58. doi: 10.1093/molbev/msq017. Epub 2010 Jan 27. Mol Biol Evol. 2010. PMID: 20106904
-
Gene duplication of the eight-stranded beta-barrel OmpX produces a functional pore: a scenario for the evolution of transmembrane beta-barrels.J Mol Biol. 2007 Mar 2;366(4):1174-84. doi: 10.1016/j.jmb.2006.12.029. Epub 2006 Dec 16. J Mol Biol. 2007. PMID: 17217961
-
Transmembrane β-barrels: Evolution, folding and energetics.Biochim Biophys Acta Biomembr. 2017 Dec;1859(12):2467-2482. doi: 10.1016/j.bbamem.2017.09.020. Epub 2017 Sep 22. Biochim Biophys Acta Biomembr. 2017. PMID: 28943271 Free PMC article. Review.
-
[Research progress on the folding and membrane-insertion mechanisms of beta-barrel outer membrane protein of gram-negative bacteria--a review].Wei Sheng Wu Xue Bao. 2014 Mar 4;54(3):261-8. Wei Sheng Wu Xue Bao. 2014. PMID: 24984517 Review. Chinese.
Cited by
-
Outer membrane protein evolution.Curr Opin Struct Biol. 2021 Jun;68:122-128. doi: 10.1016/j.sbi.2021.01.002. Epub 2021 Jan 22. Curr Opin Struct Biol. 2021. PMID: 33493965 Free PMC article. Review.
-
Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel.Nat Commun. 2019 Jul 26;10(1):3358. doi: 10.1038/s41467-019-11230-9. Nat Commun. 2019. PMID: 31350400 Free PMC article.
-
Evolutionary engineering a larger porin using a loop-to-hairpin mechanism.bioRxiv [Preprint]. 2023 Sep 20:2023.06.14.544993. doi: 10.1101/2023.06.14.544993. bioRxiv. 2023. Update in: J Mol Biol. 2023 Nov 15;435(22):168292. doi: 10.1016/j.jmb.2023.168292. PMID: 37398247 Free PMC article. Updated. Preprint.
-
Small protein folds at the root of an ancient metabolic network.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7193-7199. doi: 10.1073/pnas.1914982117. Epub 2020 Mar 18. Proc Natl Acad Sci U S A. 2020. PMID: 32188785 Free PMC article.
-
Native β-barrel substrates pass through two shared intermediates during folding on the BAM complex.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2409672121. doi: 10.1073/pnas.2409672121. Epub 2024 Oct 8. Proc Natl Acad Sci U S A. 2024. PMID: 39378083 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

