A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family
- PMID: 30496313
- PMCID: PMC6264518
- DOI: 10.1371/journal.pone.0208276
A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family
Abstract
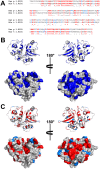
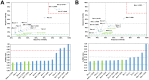
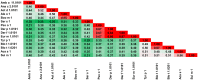
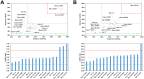
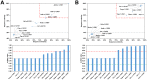
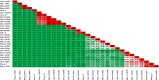
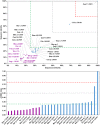
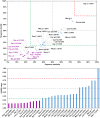
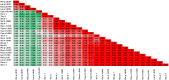
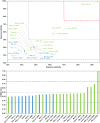
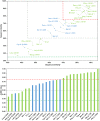
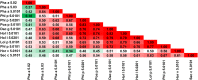
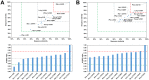
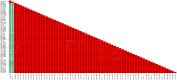
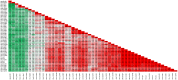
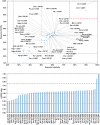
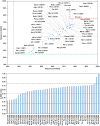
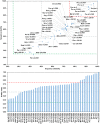
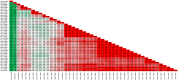
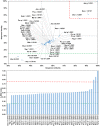
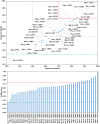
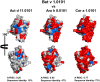
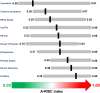
Among the vast number of identified protein families, allergens emanate from relatively few families which translates to only a small fraction of identified protein families. In allergy diagnostics and immunotherapy, interactions between immunoglobulin E and allergens are crucial because the formation of an allergen-antibody complex is necessary for triggering an allergic reaction. In allergic diseases, there is a phenomenon known as cross-reactivity. Cross-reactivity describes a situation where an individual has produced antibodies against a particular allergenic protein, but said antibodies fail to discriminate between the original sensitizer and other similar proteins that usually belong to the same family. To expound the concept of cross-reactivity, this study examines ten protein families that include allergens selected specifically for the analysis of cross-reactivity. The selected allergen families had at least 13 representative proteins, overall folds that differ significantly between families, and include relevant allergens with various potencies. The selected allergens were analyzed using information on sequence similarities and identities between members of the families as well as reports on clinically relevant cross-reactivities. Based on our analysis, we propose to introduce a new A-RISC index (Allergens'-Relative Identity, Similarity and Cross-reactivity) which describes homology between two allergens belonging to the same protein family and is used to predict the likelihood of cross-reactivity between them. Information on sequence similarities and identities, as well as on the values of the proposed A-RISC index is used to introduce four categories describing a risk of a cross-reactive reaction, namely: high, medium-high, medium-low and low. The proposed approach can facilitate analysis in component-resolved allergy diagnostics, generation of avoidance guidelines for allergic individuals, and help with the design of immunotherapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
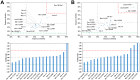
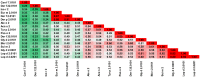
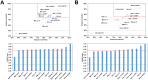
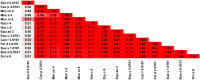


References
-
- Radauer C, Nandy A, Ferreira F, Goodman RE, Larsen JN, Lidholm J, et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy. 2014;69(4):413–9. . - PubMed
-
- Jenkins JA, Breiteneder H, Mills EN. Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol. 2007;120(6):1399–405. 10.1016/j.jaci.2007.08.019 . - DOI - PubMed
-
- Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J Allergy Clin Immunol. 2006;117(1):141–7. 10.1016/j.jaci.2005.09.010 . - DOI - PubMed
-
- Radauer C, Breiteneder H. Evolutionary biology of plant food allergens. J Allergy Clin Immunol. 2007;120(3):518–25. 10.1016/j.jaci.2007.07.024 . - DOI - PubMed
-
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol. 2008;121(4):847–52 e7. 10.1016/j.jaci.2008.01.025 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

