Developmental Xist induction is mediated by enhanced splicing
- PMID: 30496473
- PMCID: PMC6379716
- DOI: 10.1093/nar/gky1198
Developmental Xist induction is mediated by enhanced splicing
Abstract
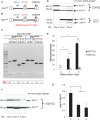
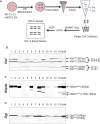
X-inactive-specific transcript (Xist) is a long noncoding RNA (lncRNA) essential for inactivating one of the two X chromosomes in mammalian females. Random X chromosome inactivation is mediated by Xist RNA expressed from the inactive X chromosome. We found that Xist RNA is unspliced in naïve embryonic stem (ES) cells. Upon differentiation, Xist splicing becomes efficient across all exons independent of transcription, suggesting interdependent or coordinated removal of Xist introns. In female cells with mutated polypyrimidine tract binding protein 1 (Ptbp1), differentiation fails to substantially upregulate mature Xist RNA because of a defect in Xist splicing. We further found both Xist129 and XistCAS RNA are unspliced in Mus musculus 129SvJ/Mus castaneous (CAS) hybrid female ES cells. Upon differentiation, Xist129 exhibits a higher splicing efficiency than XistCAS, likely contributing to preferential inhibition of the X129 chromosome. Single cell analysis shows that the allelic choice of Xist splicing is linked to the inactive X chromosome. We conclude post-transcriptional control of Xist RNA splicing is an essential regulatory step of Xist induction. Our studies shed light on the developmental roles of splicing for nuclear-retained Xist lncRNA and suggest inefficient Xist splicing is an additional fail-safe mechanism to prevent Xist activity in ES cells.
© The Author(s) 2018. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Huynh K.D., Lee J.T.. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003; 426:857–862. - PubMed
-
- Mak W., Nesterova T.B., de Napoles M., Appanah R., Yamanaka S., Otte A.P., Brockdorff N.. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004; 303:666–669. - PubMed
-
- Monkhorst K., Jonkers I., Rentmeester E., Grosveld F., Gribnau J.. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008; 132:410–421. - PubMed
-
- Lessing D., Anguera M.C., Lee J.T.. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 2013; 14:85–110. - PubMed
-
- Augui S., Nora E.P., Heard E.. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011; 12:429–442. - PubMed

