Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1
- PMID: 30496486
- PMCID: PMC6379679
- DOI: 10.1093/nar/gky1207
Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1
Abstract
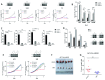
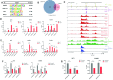
As the second most common malignant bone tumor in children and adolescents, Ewing sarcoma is initiated and exacerbated by a chimeric oncoprotein, most commonly, EWS-FLI1. In this study, we apply epigenomic analysis to characterize the transcription dysregulation in this cancer, focusing on the investigation of super-enhancer and its associated transcriptional regulatory mechanisms. We demonstrate that super-enhancer-associated transcripts are significantly enriched in EWS-FLI1 target genes, contribute to the aberrant transcriptional network of the disease, and mediate the exceptional sensitivity of Ewing sarcoma to transcriptional inhibition. Through integrative analysis, we identify MEIS1 as a super-enhancer-driven oncogene, which co-operates with EWS-FLI1 in transcriptional regulation, and plays a key pro-survival role in Ewing sarcoma. Moreover, APCDD1, another super-enhancer-associated gene, acting as a downstream target of both MEIS1 and EWS-FLI1, is also characterized as a novel tumor-promoting factor in this malignancy. These data delineate super-enhancer-mediated transcriptional deregulation in Ewing sarcoma, and uncover numerous candidate oncogenes which can be exploited for further understanding of the molecular pathogenesis for this disease.
© The Author(s) 2018. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

