Homologous Recombination and the Formation of Complex Genomic Rearrangements
- PMID: 30497856
- PMCID: PMC6402879
- DOI: 10.1016/j.tcb.2018.10.006
Homologous Recombination and the Formation of Complex Genomic Rearrangements
Abstract
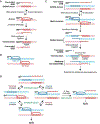
The maintenance of genome integrity involves multiple independent DNA damage avoidance and repair mechanisms. However, the origin and pathways of the focal chromosomal reshuffling phenomena collectively referred to as chromothripsis remain mechanistically obscure. We discuss here the role, mechanisms, and regulation of homologous recombination (HR) in the formation of simple and complex chromosomal rearrangements. We emphasize features of the recently characterized multi-invasion (MI)-induced rearrangement (MIR) pathway which uniquely amplifies the initial DNA damage. HR intermediates and cellular contexts that endanger genomic stability are discussed as well as the emerging roles of various classes of nucleases in the formation of genome rearrangements. Long-read sequencing and improved mapping of repeats should enable better appreciation of the significance of recombination in generating genomic rearrangements.
Keywords: chromothripsis; copynumber variation; multi-invasion; non-allelic homologous recombination; structural variant; structure-selective endonuclease.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures