Species Richness, rRNA Gene Abundance, and Seasonal Dynamics of Airborne Plant-Pathogenic Oomycetes
- PMID: 30498479
- PMCID: PMC6249755
- DOI: 10.3389/fmicb.2018.02673
Species Richness, rRNA Gene Abundance, and Seasonal Dynamics of Airborne Plant-Pathogenic Oomycetes
Erratum in
-
Corrigendum: Species Richness, rRNA Gene Abundance, and Seasonal Dynamics of Airborne Plant-Pathogenic Oomycetes.Front Microbiol. 2019 Oct 16;10:2200. doi: 10.3389/fmicb.2019.02200. eCollection 2019. Front Microbiol. 2019. PMID: 31681181 Free PMC article.
Abstract
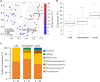
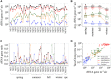
Oomycetes, also named Peronosporomycetes, are one of the most important and widespread groups of plant pathogens, leading to significant losses in the global agricultural productivity. They have been studied extensively in ground water, soil, and host plants, but their atmospheric transport vector is not well characterized. In this study, the occurrence of airborne Oomycetes was investigated by Sanger sequencing and quantitative PCR of coarse and fine aerosol particle samples (57 filter pairs) collected over a 1-year period (2006-2007) and full seasonal cycle in Mainz, Germany. In coarse particulate matter, we found 55 different hypothetical species (OTUs), of which 54 were plant pathogens and 29 belonged to the genus Peronospora (downy mildews). In fine particulate matter (<3 μm), only one species of Hyaloperonospora was found in one sample. Principal coordinate analysis of the species composition revealed three community clusters with a dependence on ambient temperature. The abundance of Oomycetes rRNA genes was low in winter and enhanced during spring, summer, and fall, with a dominance of Phytophthora, reaching a maximum concentration of ∼1.6 × 106 rRNA genes per cubic meter of sampled air in summer. The presence and high concentration of rRNA genes in air suggests that atmospheric transport, which can lead to secondary infection, may be more important than currently estimated. This illustrates the need for more current and detailed datasets, as potential seasonal shifts due to changing meteorological conditions may influence the composition of airborne Oomycetes. An insight into the dynamics of airborne plant pathogens and their major drivers should be useful for improved forecasting and management of related plant diseases.
Keywords: Peronosporomycetes; Sanger sequencing; airborne Oomycetes; meteorological parameter; plant pathogen; qPCR analysis; seasonal distribution.
Figures

References
-
- Abraham V., Kushalappa A. C., Carisse O., Bourgeois G., Auclair P. (1995). Comparison of decision methods to initiate fungicide applications against cercospora blight of carrot. Phytoprotection 76 91–99. 10.7202/706088ar - DOI
-
- Aylor D. E., Taylor G. S., Raynor G. S. (1982). Long-range transport of tobacco blue mold spores. Agric. Meteorol. 27 217–232. 10.1016/0002-1571(82)90007-3 - DOI
LinkOut - more resources
Full Text Sources

