Characterization of vaginal microbiota in Thai women
- PMID: 30498641
- PMCID: PMC6252066
- DOI: 10.7717/peerj.5977
Characterization of vaginal microbiota in Thai women
Abstract
Background: The vaginal microbiota (VMB) plays a key role in women's reproductive health. VMB composition varies with ethnicity, making it necessary to characterize the VMB of the target population before interventions to maintain and/or improve the vaginal health are undertaken. Information on the VMB of Thai women is currently unavailable. We therefore characterized the VMB in normal Thai women.
Methods: Vaginal samples derived from 25 Thai women were subjected to 16S rRNA gene next-generation sequencing (NGS) on the Ion Torrent PGM platform.
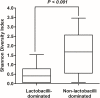
Results: Two groups of VMB were detected, lactobacilli-dominated (LD) and non-lactobacilli dominated (NLD) groups. Lactobacillus iners was the most common species found in the LD group while Gardnerella vaginalis followed by Atopobium vaginae and Pseudumonas stutzeri were commonly found in the NLD group.
Conclusions: The VMB patterns present in normal Thai women is essential information to further determine the factors associated with VMB patterns in vaginal health and disease and to develop proper management of reproductive health of Thai women.
Keywords: 16S rRNA gene next-generation sequencing; Bacterial vaginosis; Gardnerella vaginalis; Lactobacillus; Thailand; Vaginal microbiota.
Conflict of interest statement
The authors declare there are no competing interests.
Figures





References
-
- Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, Gevers D, Huttenhower C, Petrosino J, Versalovic J. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLOS ONE. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. - DOI - PMC - PubMed
-
- Borgdorff H, Van der Veer C, Van Houdt R, Alberts CJ, De Vries HJ, Bruisten SM, Snijder MB, Prins M, Geerlings SE, Schim van der Loeff MF, Van de Wijgert J. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLOS ONE. 2017;12:e0181135. doi: 10.1371/journal.pone.0181135. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

