Danger signals activate a putative innate immune system during regeneration in a filamentous fungus
- PMID: 30500812
- PMCID: PMC6291166
- DOI: 10.1371/journal.pgen.1007390
Danger signals activate a putative innate immune system during regeneration in a filamentous fungus
Abstract
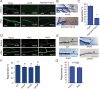
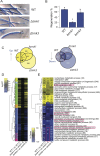
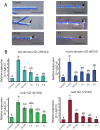
The ability to respond to injury is a biological process shared by organisms of different kingdoms that can even result in complete regeneration of a part or structure that was lost. Due to their immobility, multicellular fungi are prey to various predators and are therefore constantly exposed to mechanical damage. Nevertheless, our current knowledge of how fungi respond to injury is scarce. Here we show that activation of injury responses and hyphal regeneration in the filamentous fungus Trichoderma atroviride relies on the detection of two danger or alarm signals. As an early response to injury, we detected a transient increase in cytosolic free calcium ([Ca2+]c) that was promoted by extracellular ATP, and which is likely regulated by a mechanism of calcium-induced calcium-release. In addition, we demonstrate that the mitogen activated protein kinase Tmk1 plays a key role in hyphal regeneration. Calcium- and Tmk1-mediated signaling cascades activated major transcriptional changes early following injury, including induction of a set of regeneration associated genes related to cell signaling, stress responses, transcription regulation, ribosome biogenesis/translation, replication and DNA repair. Interestingly, we uncovered the activation of a putative fungal innate immune response, including the involvement of HET domain genes, known to participate in programmed cell death. Our work shows that fungi and animals share danger-signals, signaling cascades, and the activation of the expression of genes related to immunity after injury, which are likely the result of convergent evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Sanchez Alvarado A. Regeneration in the metazoans: why does it happen? Bioessays 2000; 22: 578–590. 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-# - DOI - PubMed
-
- Heil M, Land WG. Danger signals-damaged-self recognition across the tree of life. Front. Plant Sci. 2014; 5: 578 10.3389/fpls.2014.00578 - DOI - PMC - PubMed
-
- Atwal JK, Massie B, Miller FD, Kaplan DR. The TrkB-Shc site signals neuronal survival and local axon growth via MEK and P13-kinase. Neuron 2000; 27: 265–277. - PubMed
-
- Hettenhausen C, Schuman MC, Wu J. MAPK signaling: a key element in plant defense response to insects. Insect Sci. 2015; 22: 157–164. 10.1111/1744-7917.12128 - DOI - PMC - PubMed
-
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, et al. Identification of a plant receptor for extracellular ATP. Science 2014; 343: 290–294. 10.1126/science.343.6168.290 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

