De novo mutations in the GTP/GDP-binding region of RALA, a RAS-like small GTPase, cause intellectual disability and developmental delay
- PMID: 30500825
- PMCID: PMC6291162
- DOI: 10.1371/journal.pgen.1007671
De novo mutations in the GTP/GDP-binding region of RALA, a RAS-like small GTPase, cause intellectual disability and developmental delay
Abstract
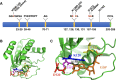
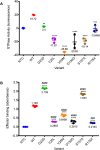
Mutations that alter signaling of RAS/MAPK-family proteins give rise to a group of Mendelian diseases known as RASopathies. However, among RASopathies, the matrix of genotype-phenotype relationships is still incomplete, in part because there are many RAS-related proteins and in part because the phenotypic consequences may be variable and/or pleiotropic. Here, we describe a cohort of ten cases, drawn from six clinical sites and over 16,000 sequenced probands, with de novo protein-altering variation in RALA, a RAS-like small GTPase. All probands present with speech and motor delays, and most have intellectual disability, low weight, short stature, and facial dysmorphism. The observed rate of de novo RALA variants in affected probands is significantly higher (p = 4.93 x 10(-11)) than expected from the estimated random mutation rate. Further, all de novo variants described here affect residues within the GTP/GDP-binding region of RALA; in fact, six alleles arose at only two codons, Val25 and Lys128. The affected residues are highly conserved across both RAL- and RAS-family genes, are devoid of variation in large human population datasets, and several are homologous to positions at which disease-associated variants have been observed in other GTPase genes. We directly assayed GTP hydrolysis and RALA effector-protein binding of the observed variants, and found that all but one tested variant significantly reduced both activities compared to wild-type. The one exception, S157A, reduced GTP hydrolysis but significantly increased RALA-effector binding, an observation similar to that seen for oncogenic RAS variants. These results show the power of data sharing for the interpretation and analysis of rare variation, expand the spectrum of molecular causes of developmental disability to include RALA, and provide additional insight into the pathogenesis of human disease caused by mutations in small GTPases.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: GMC is currently serving as an Academic Editor for PLOS Genetics. GSB is currently serving as an Editor-In-Chief for PLOS Genetics. ZP is an employee of Ambry Genetics, which provides exome sequencing as a commercially available test. IMW, RW, SFS are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. that also offers commercial exome sequencing. The remaining authors declare no conflicts of interest.
Figures

Comment in
-
Emerging RAS superfamily conditions involving GTPase function.PLoS Genet. 2019 Feb 14;15(2):e1007870. doi: 10.1371/journal.pgen.1007870. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30763311 Free PMC article. No abstract available.
References
-
- Ropers HH. Genetics of intellectual disability. Curr Opin Genet Dev. 2008;18(3):241–50. 10.1016/j.gde.2008.07.008 - DOI - PubMed
-
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–9. 10.1001/jama.2014.14601 - DOI - PMC - PubMed
-
- Taylor JC, Martin HC, Lise S, Broxholme J, Cazier JB, Rimmer A, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47(7):717–26. 10.1038/ng.3304 - DOI - PMC - PubMed
-
- Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170(1):17–33. 10.1016/j.cell.2017.06.009 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

