Nephromyces Encodes a Urate Metabolism Pathway and Predicted Peroxisomes, Demonstrating That These Are Not Ancient Losses of Apicomplexans
- PMID: 30500900
- PMCID: PMC6320678
- DOI: 10.1093/gbe/evy251
Nephromyces Encodes a Urate Metabolism Pathway and Predicted Peroxisomes, Demonstrating That These Are Not Ancient Losses of Apicomplexans
Abstract
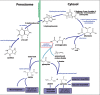
The phylum Apicomplexa is a quintessentially parasitic lineage, whose members infect a broad range of animals. One exception to this may be the apicomplexan genus Nephromyces, which has been described as having a mutualistic relationship with its host. Here we analyze transcriptome data from Nephromyces and its parasitic sister taxon, Cardiosporidium, revealing an ancestral purine degradation pathway thought to have been lost early in apicomplexan evolution. The predicted localization of many of the purine degradation enzymes to peroxisomes, and the in silico identification of a full set of peroxisome proteins, indicates that loss of both features in other apicomplexans occurred multiple times. The degradation of purines is thought to play a key role in the unusual relationship between Nephromyces and its host. Transcriptome data confirm previous biochemical results of a functional pathway for the utilization of uric acid as a primary nitrogen source for this unusual apicomplexan.
Figures

Similar articles
-
Nephromyces Represents a Diverse and Novel Lineage of the Apicomplexa That Has Retained Apicoplasts.Genome Biol Evol. 2019 Oct 1;11(10):2727-2740. doi: 10.1093/gbe/evz155. Genome Biol Evol. 2019. PMID: 31328784 Free PMC article.
-
Metabolic Contributions of an Alphaproteobacterial Endosymbiont in the Apicomplexan Cardiosporidium cionae.Front Microbiol. 2020 Dec 1;11:580719. doi: 10.3389/fmicb.2020.580719. eCollection 2020. Front Microbiol. 2020. PMID: 33335517 Free PMC article.
-
Nephromyces, a beneficial apicomplexan symbiont in marine animals.Proc Natl Acad Sci U S A. 2010 Sep 14;107(37):16190-5. doi: 10.1073/pnas.1002335107. Epub 2010 Aug 24. Proc Natl Acad Sci U S A. 2010. PMID: 20736348 Free PMC article.
-
Evolution of urate-degrading enzymes in animal peroxisomes.Cell Biochem Biophys. 2000;32 Spring:123-9. doi: 10.1385/cbb:32:1-3:123. Cell Biochem Biophys. 2000. PMID: 11330038 Review.
-
Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: insights from comparative genomics.Int Rev Cytol. 2007;262:1-74. doi: 10.1016/S0074-7696(07)62001-4. Int Rev Cytol. 2007. PMID: 17631186 Review.
Cited by
-
Nephromyces Represents a Diverse and Novel Lineage of the Apicomplexa That Has Retained Apicoplasts.Genome Biol Evol. 2019 Oct 1;11(10):2727-2740. doi: 10.1093/gbe/evz155. Genome Biol Evol. 2019. PMID: 31328784 Free PMC article.
-
Adaptations and metabolic evolution of myzozoan protists across diverse lifestyles and environments.Microbiol Mol Biol Rev. 2024 Dec 18;88(4):e0019722. doi: 10.1128/mmbr.00197-22. Epub 2024 Oct 10. Microbiol Mol Biol Rev. 2024. PMID: 39387588 Review.
-
Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential.Mar Drugs. 2021 Jun 26;19(7):370. doi: 10.3390/md19070370. Mar Drugs. 2021. PMID: 34206769 Free PMC article. Review.
-
Metabolic Contributions of an Alphaproteobacterial Endosymbiont in the Apicomplexan Cardiosporidium cionae.Front Microbiol. 2020 Dec 1;11:580719. doi: 10.3389/fmicb.2020.580719. eCollection 2020. Front Microbiol. 2020. PMID: 33335517 Free PMC article.
-
Inferred Subcellular Localization of Peroxisomal Matrix Proteins of Guillardia theta Suggests an Important Role of Peroxisomes in Cryptophytes.Front Plant Sci. 2022 Jun 16;13:889662. doi: 10.3389/fpls.2022.889662. eCollection 2022. Front Plant Sci. 2022. PMID: 35783940 Free PMC article.
References
-
- Arrowood MJ, Sterling CR.. 1987. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol. 73(2):314–319. - PubMed
-
- Cardoso R, Soares H, Hemphill A, Leitão A.. 2016. Apicomplexans pulling the strings: manipulation of the host cell cytoskeleton dynamics. Parasitology 143(8):957–970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

