Tales of Detailed Poly(A) Tails
- PMID: 30503240
- PMCID: PMC7083203
- DOI: 10.1016/j.tcb.2018.11.002
Tales of Detailed Poly(A) Tails
Abstract
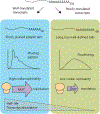
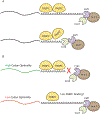
Poly(A) tails are non-templated additions of adenosines at the 3' ends of most eukaryotic mRNAs. In the nucleus, these RNAs are co-transcriptionally cleaved at a poly(A) site and then polyadenylated before being exported to the cytoplasm. In the cytoplasm, poly(A) tails play pivotal roles in the translation and stability of the mRNA. One challenge in studying poly(A) tails is that they are difficult to sequence and accurately measure. However, recent advances in sequencing technology, computational algorithms, and other assays have enabled a more detailed look at poly(A) tail length genome-wide throughout many developmental stages and organisms. With the help of these advances, our understanding of poly(A) tail length has evolved over the past 5 years with the recognition that highly expressed genes can have short poly(A) tails and the elucidation of the seemingly contradictory roles for poly(A)-binding protein (PABP) in facilitating both protection and deadenylation.
Keywords: deadenylation; poly(A) tail; poly(A)-binding protein; translation.
Copyright © 2018. Published by Elsevier Ltd.
Figures

References
-
- Goldstrohm AC and Wickens M (2008) Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol 9, 337–344 - PubMed
-
- Schmidt MJ and Norbury CJ (2010) Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip. Rev. RNA 1, 142–151 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

