Temperature Sensing Is Distributed throughout the Regulatory Network that Controls FLC Epigenetic Silencing in Vernalization
- PMID: 30503646
- PMCID: PMC6310686
- DOI: 10.1016/j.cels.2018.10.011
Temperature Sensing Is Distributed throughout the Regulatory Network that Controls FLC Epigenetic Silencing in Vernalization
Abstract
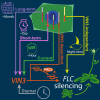
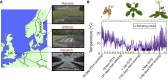
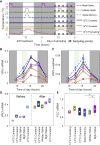
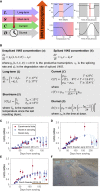
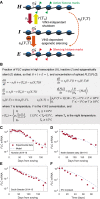
Many organisms need to respond to complex, noisy environmental signals for developmental decision making. Here, we dissect how Arabidopsis plants integrate widely fluctuating field temperatures over month-long timescales to progressively upregulate VERNALIZATION INSENSITIVE3 (VIN3) and silence FLOWERING LOCUS C (FLC), aligning flowering with spring. We develop a mathematical model for vernalization that operates on multiple timescales-long term (month), short term (day), and current (hour)-and is constrained by experimental data. Our analysis demonstrates that temperature sensing is not localized to specific nodes within the FLC network. Instead, temperature sensing is broadly distributed, with each thermosensory process responding to specific features of the plants' history of exposure to warm and cold. The model accurately predicts FLC silencing in new field data, allowing us to forecast FLC expression in changing climates. We suggest that distributed thermosensing may be a general property of thermoresponsive regulatory networks in complex natural environments.
Keywords: FLC; FLOWERING LOCUS C; VERNALIZATION INSENSITIVE3; VIN3; climate change; epigenetics; gene regulation; mathematical modeling; phenology; temperature sensing; vernalization.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- BBS/E/J/000PR9773/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000PR9788/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004588/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P013511/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

