Global analysis of osteosarcoma lipidomes reveal altered lipid profiles in metastatic versus nonmetastatic cells
- PMID: 30504231
- PMCID: PMC6358301
- DOI: 10.1194/jlr.M088559
Global analysis of osteosarcoma lipidomes reveal altered lipid profiles in metastatic versus nonmetastatic cells
Abstract
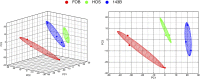
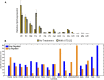
Osteosarcoma (OS) is the most common form of primary bone cancer in humans. The early detection and subsequent control of metastasis has been challenging in OS. Lipids are important constituents of cells that maintain structural integrity that can be converted into lipid-signaling molecules and are reprogrammed in cancerous states. Here, we investigate the global lipidomic differences in metastatic (143B) and nonmetastatic (HOS) human OS cells as compared with normal fetal osteoblast cells (FOB) using lipidomics. We detect 15 distinct lipid classes in all three cell lines that included over 1,000 lipid species across various classes including phospholipids, sphingolipids and ceramides, glycolipids, and cholesterol. We identify a key class of lipids, diacylglycerols, which are overexpressed in metastatic OS cells as compared with their nonmetastatic or nontumorigenic counterparts. As a proof of concept, we show that blocking diacylglycerol synthesis reduces cellular viability and reduces cell migration in metastatic OS cells. Thus, the differentially regulated lipids identified in this study might aid in biomarker discovery, and the synthesis and metabolism of specific lipids could serve as future targets for therapeutic development.
Keywords: cholesterol; diacylglycerol; lipidomics; mass spectrometry.
Copyright © 2019 Roy et al.
Figures






References
-
- Bielack S. S., Kempf-Bielack B., Delling G., Exner G. U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., et al. 2002. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 20: 776–790. - PubMed
-
- Dopico A. M., and Tigyi G. J.. 2007. A glance at the structural and functional diversity of membrane lipids. Methods Mol. Biol. 400: 1–13. - PubMed
-
- Shevchenko A., and Simons K.. 2010. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11: 593–598. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

