RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage
- PMID: 30504444
- PMCID: PMC6352405
- DOI: 10.1136/annrheumdis-2018-213882
RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage
Abstract
Objective: To uncover the microRNA (miRNA) interactome of the osteoarthritis (OA) pathophysiological process in the cartilage.
Methods: We performed RNA sequencing in 130 samples (n=35 and n=30 pairs for messenger RNA (mRNA) and miRNA, respectively) on macroscopically preserved and lesioned OA cartilage from the same patient and performed differential expression (DE) analysis of miRNA and mRNAs. To build an OA-specific miRNA interactome, a prioritisation scheme was applied based on inverse Pearson's correlations and inverse DE of miRNAs and mRNAs. Subsequently, these were filtered by those present in predicted (TargetScan/microT-CDS) and/or experimentally validated (miRTarBase/TarBase) public databases. Pathway enrichment analysis was applied to elucidate OA-related pathways likely mediated by miRNA regulatory mechanisms.
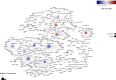
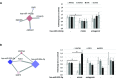
Results: We found 142 miRNAs and 2387 mRNAs to be differentially expressed between lesioned and preserved OA articular cartilage. After applying prioritisation towards likely miRNA-mRNA targets, a regulatory network of 62 miRNAs targeting 238 mRNAs was created. Subsequent pathway enrichment analysis of these mRNAs (or genes) elucidated that genes within the 'nervous system development' are likely mediated by miRNA regulatory mechanisms (familywise error=8.4×10-5). Herein NTF3 encodes neurotrophin-3, which controls survival and differentiation of neurons and which is closely related to the nerve growth factor.
Conclusions: By an integrated approach of miRNA and mRNA sequencing data of OA cartilage, an OA miRNA interactome and related pathways were elucidated. Our functional data demonstrated interacting levels at which miRNA affects expression of genes in the cartilage and exemplified the complexity of functionally validating a network of genes that may be targeted by multiple miRNAs.
Keywords: data integration; mRNA; microRNAs; osteoarthritis; regulatory networks.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

