Identification of Immune Signatures of Novel Adjuvant Formulations Using Machine Learning
- PMID: 30504893
- PMCID: PMC6269591
- DOI: 10.1038/s41598-018-35452-x
Identification of Immune Signatures of Novel Adjuvant Formulations Using Machine Learning
Abstract
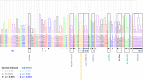
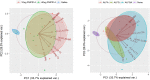
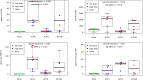
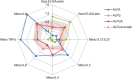
Adjuvants have long been critical components of vaccines, but the exact mechanisms of their action and precisely how they alter or enhance vaccine-induced immune responses are often unclear. In this study, we used broad immunoprofiling of antibody, cellular, and cytokine responses, combined with data integration and machine learning to gain insight into the impact of different adjuvant formulations on vaccine-induced immune responses. A Self-Assembling Protein Nanoparticles (SAPN) presenting the malarial circumsporozoite protein (CSP) was used as a model vaccine, adjuvanted with three different liposomal formulations: liposome plus Alum (ALFA), liposome plus QS21 (ALFQ), and both (ALFQA). Using a computational approach to integrate the immunoprofiling data, we identified distinct vaccine-induced immune responses and developed a multivariate model that could predict the adjuvant condition from immune response data alone with 92% accuracy (p = 0.003). The data integration also revealed that commonly used readouts (i.e. serology, frequency of T cells producing IFN-γ, IL2, TNFα) missed important differences between adjuvants. In summary, broad immune-profiling in combination with machine learning methods enabled the reliable and clear definition of immune signatures for different adjuvant formulations, providing a means for quantitatively characterizing the complex roles that adjuvants can play in vaccine-induced immunity. The approach described here provides a powerful tool for identifying potential immune correlates of protection, a prerequisite for the rational pairing of vaccines candidates and adjuvants.
Conflict of interest statement
Dr. Kevin Beck is an employee of Miltenyi Biotec Inc. (San Diego, CA).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

