Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential
- PMID: 30504906
- PMCID: PMC6269548
- DOI: 10.1038/s41467-018-07631-x
Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential
Abstract
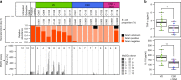
The rate of caesarean section delivery (CSD) is increasing worldwide. It remains unclear whether disruption of mother-to-neonate transmission of microbiota through CSD occurs and whether it affects human physiology. Here we perform metagenomic analysis of earliest gut microbial community structures and functions. We identify differences in encoded functions between microbiomes of vaginally delivered (VD) and CSD neonates. Several functional pathways are over-represented in VD neonates, including lipopolysaccharide (LPS) biosynthesis. We link these enriched functions to individual-specific strains, which are transmitted from mothers to neonates in case of VD. The stimulation of primary human immune cells with LPS isolated from early stool samples of VD neonates results in higher levels of tumour necrosis factor (TNF-α) and interleukin 18 (IL-18). Accordingly, the observed levels of TNF-α and IL-18 in neonatal blood plasma are higher after VD. Taken together, our results support that CSD disrupts mother-to-neonate transmission of specific microbial strains, linked functional repertoires and immune-stimulatory potential during a critical window for neonatal immune system priming.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

