Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections
- PMID: 30504915
- PMCID: PMC6269475
- DOI: 10.1038/s41467-018-07570-7
Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections
Abstract
The increasing number of severe infections with multi-drug-resistant pathogens worldwide highlights the need for alternative treatment options. Given the pivotal role of phagocytes and especially alveolar macrophages in pulmonary immunity, we introduce a new, cell-based treatment strategy to target bacterial airway infections. Here we show that the mass production of therapeutic phagocytes from induced pluripotent stem cells (iPSC) in industry-compatible, stirred-tank bioreactors is feasible. Bioreactor-derived iPSC-macrophages (iPSC-Mac) represent a highly pure population of CD45+CD11b+CD14+CD163+ cells, and share important phenotypic, functional and transcriptional hallmarks with professional phagocytes, however with a distinct transcriptome signature similar to primitive macrophages. Most importantly, bioreactor-derived iPSC-Mac rescue mice from Pseudomonas aeruginosa-mediated acute infections of the lower respiratory tract within 4-8 h post intra-pulmonary transplantation and reduce bacterial load. Generation of specific immune-cells from iPSC-sources in scalable stirred-tank bioreactors can extend the field of immunotherapy towards bacterial infections, and may allow for further innovative cell-based treatment strategies.
Conflict of interest statement
Part of this work is included in a patent application. M.A., H.K., T.M., R.Z., and N.L. are authors of the patent application (European patent application number PCT/EP2018/061574) entitled “Stem-cell derived myeloid cells, generation and use thereof”. The priority date of the application is 04.05.2017. All the remaining authors declare no competiting interests.
Figures
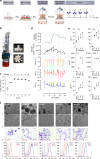
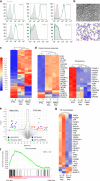
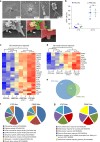

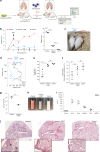
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

