Utilization of a Strongly Inducible DDI2 Promoter to Control Gene Expression in Saccharomyces cerevisiae
- PMID: 30505295
- PMCID: PMC6250804
- DOI: 10.3389/fmicb.2018.02736
Utilization of a Strongly Inducible DDI2 Promoter to Control Gene Expression in Saccharomyces cerevisiae
Abstract
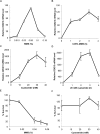
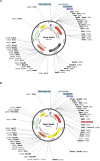
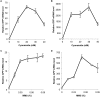
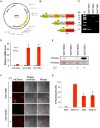
Regulating target gene expression is a common method in yeast research. In Saccharomyces cerevisiae, there are several widely used regulated expression systems, such as the GAL and Tet-off systems. However, all current expression systems possess some intrinsic deficiencies. We have previously reported that the DDI2 gene can be induced to very high levels upon cyanamide or methyl methanesulfonate treatment. Here we report the construction of gene expression systems based on the DDI2 promoter in both single- and multi-copy plasmids. Using GFP as a reporter gene, it was demonstrated that the target gene expression could be increased by up to 2,000-fold at the transcriptional level by utilizing the above systems. In addition, a DDI2-based construct was created for promoter shuffling in the budding yeast genome to control endogenous gene expression. Overall, this study offers a set of convenient and highly efficient experimental tools to control target gene expression in budding yeast.
Keywords: DDI2; Saccharomyces cerevisiae; cyanamide; promoter; transcriptional regulation.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

