Functional Heterogeneity of CD4+ Tumor-Infiltrating Lymphocytes With a Resident Memory Phenotype in NSCLC
- PMID: 30505306
- PMCID: PMC6250821
- DOI: 10.3389/fimmu.2018.02654
Functional Heterogeneity of CD4+ Tumor-Infiltrating Lymphocytes With a Resident Memory Phenotype in NSCLC
Abstract
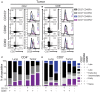
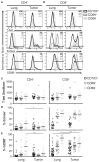
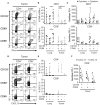
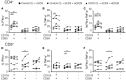
Resident memory T cells (TRM) inhabit peripheral tissues and are critical for protection against localized infections. Recently, it has become evident that CD103+ TRM are not only important in combating secondary infections, but also for the elimination of tumor cells. In several solid cancers, intratumoral CD103+CD8+ tumor infiltrating lymphocytes (TILs), with TRM properties, are a positive prognostic marker. To better understand the role of TRM in tumors, we performed a detailed characterization of CD8+ and CD4+ TIL phenotype and functional properties in non-small cell lung cancer (NSCLC). Frequencies of CD8+ and CD4+ T cell infiltrates in tumors were comparable, but we observed a sharp contrast in TRM ratios compared to surrounding lung tissue. The majority of both CD4+ and CD8+ TILs expressed CD69 and a subset also expressed CD103, both hallmarks of TRM. While CD103+CD8+ T cells were enriched in tumors, CD103+CD4+ T cell frequencies were decreased compared to surrounding lung tissue. Furthermore, CD103+CD4+ and CD103+CD8+ TILs showed multiple characteristics of TRM, such as elevated expression of CXCR6 and CD49a, and decreased expression of T-bet and Eomes. In line with the immunomodulatory role of the tumor microenvironment, CD8+ and CD4+ TILs expressed high levels of inhibitory receptors 2B4, CTLA-4, and PD-1, with the highest levels found on CD103+ TILs. Strikingly, CD103+CD4+ TILs were the most potent producers of TNF-α and IFN-γ, while other TIL subsets lacked such cytokine production. Whereas, CD103+CD4+PD-1low TILs produced the most effector cytokines, CD103+CD4+PD-1++ and CD69+CD4+PD-1++ TILs produced CXCL13. Furthermore, a large proportion of TILs expressed co-stimulatory receptors CD27 and CD28, unlike lung TRM, suggesting a less differentiated phenotype. Agonistic triggering of these receptors improved cytokine production of CD103+CD4+ and CD69+CD8+ TILs. Our findings thus provide a rationale to target CD103+CD4+ TILs and add co-stimulation to current therapies to improve the efficacy of immunotherapies and cancer vaccines.
Keywords: NSCLC; TILs; TRM; co-stimulation; cytokines; differentiation; exhaustion.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

