Mapping a diversity of genetic interactions in yeast
- PMID: 30505984
- PMCID: PMC6269142
- DOI: 10.1016/j.coisb.2017.08.002
Mapping a diversity of genetic interactions in yeast
Abstract
Genetic interactions occur when the combination of multiple mutations yields an unexpected phenotype, and they may confound our ability to fully understand the genetic mechanisms underlying complex diseases. Genetic interactions are challenging to study because there are millions of possible different variant combinations within a given genome. Consequently, they have primarily been systematically explored in unicellular model organisms, such as yeast, with a focus on pairwise genetic interactions between loss-of-function alleles. However, there are many different types of genetic interactions, such as those occurring between gain-of-function or heterozygous mutations. Here, we review recent advances made in the systematic analysis of such diverse genetic interactions in yeast, and briefly discuss how similar studies could be undertaken in human cells.
Keywords: complex haploinsufficiency; dosage interactions; epistasis; genetic interactions; higher-order interactions.
Figures
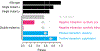
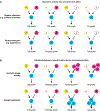

References
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al.: Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418:387–391. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
