A stochastic framework to model axon interactions within growing neuronal populations
- PMID: 30507939
- PMCID: PMC6292646
- DOI: 10.1371/journal.pcbi.1006627
A stochastic framework to model axon interactions within growing neuronal populations
Abstract
The confined and crowded environment of developing brains imposes spatial constraints on neuronal cells that have evolved individual and collective strategies to optimize their growth. These include organizing neurons into populations extending their axons to common target territories. How individual axons interact with each other within such populations to optimize innervation is currently unclear and difficult to analyze experimentally in vivo. Here, we developed a stochastic model of 3D axon growth that takes into account spatial environmental constraints, physical interactions between neighboring axons, and branch formation. This general, predictive and robust model, when fed with parameters estimated on real neurons from the Drosophila brain, enabled the study of the mechanistic principles underlying the growth of axonal populations. First, it provided a novel explanation for the diversity of growth and branching patterns observed in vivo within populations of genetically identical neurons. Second, it uncovered that axon branching could be a strategy optimizing the overall growth of axons competing with others in contexts of high axonal density. The flexibility of this framework will make it possible to investigate the rules underlying axon growth and regeneration in the context of various neuronal populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





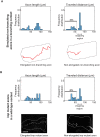
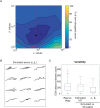
References
-
- Bixby JL, Harris WA. Molecular mechanisms of axon growth and guidance. Annual Review of Cell Biology. 1991;7:117–59. 10.1146/annurev.cb.07.110191.001001 - DOI - PubMed
-
- Tessier-Lavigne M, Goodman CS, et al. The molecular biology of axon guidance. Science. 1996;274(5290):1123–1133. 10.1126/science.274.5290.1123 - DOI - PubMed
-
- Song Hj, Poo Mm. The cell biology of neuronal navigation. Nature cell biology. 2001;3(3):E81–E88. 10.1038/35060164 - DOI - PubMed
-
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298(5600):1959–1964. 10.1126/science.1072165 - DOI - PubMed
-
- Guan KL, Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nature Reviews Neuroscience. 2003;4(12):941–956. 10.1038/nrn1254 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

