A comprehensive ensemble model for comparing the allosteric effect of ordered and disordered proteins
- PMID: 30507941
- PMCID: PMC6292653
- DOI: 10.1371/journal.pcbi.1006393
A comprehensive ensemble model for comparing the allosteric effect of ordered and disordered proteins
Abstract
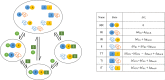
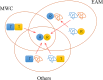
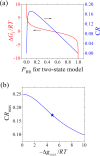
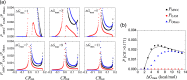
Intrinsically disordered proteins/regions (IDPs/IDRs) are prevalent in allosteric regulation. It was previously thought that intrinsic disorder is favorable for maximizing the allosteric coupling. Here, we propose a comprehensive ensemble model to compare the roles of both order-order transition and disorder-order transition in allosteric effect. It is revealed that the MWC pathway (order-order transition) has a higher probability than the EAM pathway (disorder-order transition) in allostery, suggesting a complicated role of IDPs/IDRs in regulatory proteins. In addition, an analytic formula for the maximal allosteric coupling response is obtained, which shows that too stable or too unstable state is unfavorable to endow allostery, and is thus helpful for rational design of allosteric drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Fenton AW (2008) Allostery: an illustrated definition for the 'second secret of life'. Trends In Biochemical Sciences 33: 420–425. 10.1016/j.tibs.2008.05.009 - DOI - PMC - PubMed
-
- Monod J, Changeux JP, Jacob F (1963) Allosteric proteins and cellular control systems. Journal Of Molecular Biology 6: 306–329. - PubMed
-
- Monod J, Wyman J, Changeux JP (1965) On nature of allosteric transitions—a plausible model. Journal Of Molecular Biology 12: 88–118. - PubMed
-
- Koshland DE, Nemethy G, Filmer D (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5: 365–385. - PubMed
-
- Eaton WA, Henry ER, Hofrichter J, Mozzarelli A (1999) Is cooperative oxygen binding by hemoglobin really understood? Nature Structural Biology 6: 351–358. 10.1038/7586 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

