RG108 increases NANOG and OCT4 in bone marrow-derived mesenchymal cells through global changes in DNA modifications and epigenetic activation
- PMID: 30507955
- PMCID: PMC6277091
- DOI: 10.1371/journal.pone.0207873
RG108 increases NANOG and OCT4 in bone marrow-derived mesenchymal cells through global changes in DNA modifications and epigenetic activation
Abstract
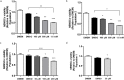
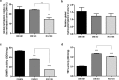
Human bone marrow-derived mesenchymal stem cells (hBMSCs) are important for tissue regeneration but their epigenetic regulation is not well understood. Here we investigate the ability of a non-nucleoside DNA methylation inhibitor, RG108 to induce epigenetic changes at both global and gene-specific levels in order to enhance mesenchymal cell markers, in hBMSCs. hBMSCs were treated with complete culture medium, 50 μM RG108 and DMSO for three days and subjected to viability and apoptosis assays, global and gene-specific methylation/hydroxymethylation, transcript levels' analysis of epigenetic machinery enzymes and multipotency markers, protein activities of DNMTs and TETs, immunofluorescence staining and western blot analysis for NANOG and OCT4 and flow cytometry for CD105. The RG108, when used at 50 μM, did not affect the viability, apoptosis and proliferation rates of hBMSCs or hydroxymethylation global levels while leading to 75% decrease in DNMTs activity and 42% loss of global DNA methylation levels. In addition, DNMT1 was significantly downregulated while TET1 was upregulated, potentially contributing to the substantial loss of methylation observed. Most importantly, the mesenchymal cell markers CD105, NANOG and OCT4 were upregulated being NANOG and OCT4 epigenetically modulated by RG108, at their gene promoters. We propose that RG108 could be used for epigenetic modulation, promoting epigenetic activation of NANOG and OCT4, without affecting the viability of hBMSCs. DMSO can be considered a modulator of epigenetic machinery enzymes, although with milder effect compared to RG108.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Greco SJ, Liu K and Rameshwar P. Functional similarities among genes regulated by OCT4 in human mesenchymal and embryonic stem cells. Stem Cells. 2007; 25: 3143–3154. 10.1634/stemcells.2007-0351 - DOI - PubMed
-
- Wei X and Shen CY. Transcriptional regulation of Oct4 in human bone marrow mesenchymal stem cells. Stem Cells Dev. 2011. 10.1089/scd.2010.0069 - DOI - PubMed
-
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122: 947–956. 10.1016/j.cell.2005.08.020 - DOI - PMC - PubMed
-
- Tsai CC, Su PF, Huang YF, Yew TL and Hung SC. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell. 2012. 10.1016/j.molcel.2012.06.020 - DOI - PubMed
-
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013. 10.1038/nature11925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

