KnotProt 2.0: a database of proteins with knots and other entangled structures
- PMID: 30508159
- PMCID: PMC6323932
- DOI: 10.1093/nar/gky1140
KnotProt 2.0: a database of proteins with knots and other entangled structures
Abstract
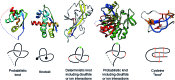
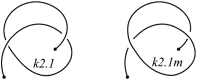
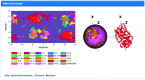
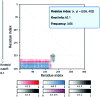
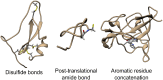
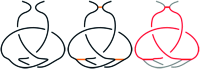
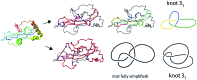
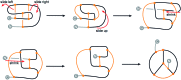
The KnotProt 2.0 database (the updated version of the KnotProt database) collects information about proteins which form knots and other entangled structures. New features in KnotProt 2.0 include the characterization of both probabilistic and deterministic entanglements which can be formed by disulfide bonds and interactions via ions, a refined characterization of entanglement in terms of knotoids, the identification of the so-called cysteine knots, the possibility to analyze all or a non-redundant set of proteins, and various technical updates. The KnotProt 2.0 database classifies all entangled proteins, represents their complexity in the form of a knotting fingerprint, and presents many biological and geometrical statistics based on these results. Currently the database contains >2000 entangled structures, and it regularly self-updates based on proteins deposited in the Protein Data Bank (PDB).
Figures




References
-
- Mansfield M.L. Are there knots in proteins. Nat. Struct. Biol. 1994; 1:213–214. - PubMed
-
- Taylor W.R. A deeply knotted protein and how it might fold. Nature. 2000; 406:916–919. - PubMed
-
- Sulkowska J.I., Sulkowski P.. Entangled proteins: knots, slipknots, links, and lassos. The Role of Topology in Materials. 2018; Springer; 201–226.

