Staphylococcus aureus toxin LukSF dissociates from its membrane receptor target to enable renewed ligand sequestration
- PMID: 30509126
- PMCID: PMC6404581
- DOI: 10.1096/fj.201801910R
Staphylococcus aureus toxin LukSF dissociates from its membrane receptor target to enable renewed ligand sequestration
Abstract
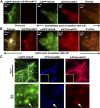
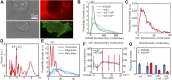
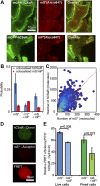
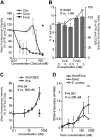
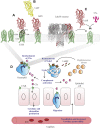
Staphylococcus aureus Panton-Valentine leukocidin is a pore-forming toxin targeting the human C5a receptor (hC5aR), enabling this pathogen to battle the immune response by destroying phagocytes through targeted lysis. The mechanisms that contribute to rapid cell lysis are largely unexplored. Here, we show that cell lysis may be enabled by a process of toxins targeting receptor clusters and present indirect evidence for receptor "recycling" that allows multiple toxin pores to be formed close together. With the use of live cell single-molecule super-resolution imaging, Förster resonance energy transfer and nanoscale total internal reflection fluorescence colocalization microscopy, we visualized toxin pore formation in the presence of its natural docking ligand. We demonstrate disassociation of hC5aR from toxin complexes and simultaneous binding of new ligands. This effect may free mobile receptors to amplify hyperinflammatory reactions in early stages of microbial infections and have implications for several other similar bicomponent toxins and the design of new antibiotics.-Haapasalo, K., Wollman, A. J. M., de Haas, C. J. C., van Kessel, K. P. M., van Strijp, J. A. G., Leake, M. C. Staphylococcus aureus toxin LukSF dissociates from its membrane receptor target to enable renewed ligand sequestration.
Keywords: bacterial toxin; immune response; pore formation; single molecule; super-resolution.
Conflict of interest statement
The authors thank Piet Aerts and Angelino Tromp (both from University Medical Center Utrecht) for assistance in sample preparation and labeling, Esther van’t Veld and Richard Wubbolts [both from Utrecht University (UU)] for assistance with light microscopy, and Dr. Karin Strijbis (UU) for providing the sortase A enzyme. This work was supported by The Finnish Cultural Foundation (Grants 00131060 and 00142390); Biological Physical Sciences Institute, Royal Society, Medical Research Council (MRC; Grant MR/K01580X/1); Biotechnology and Biological Sciences Research Council (BBSRC; Grant BB/N006453/1); Engineering and Physical Sciences Research Council (EPSRC) Physics of Life UK Network; and Wellcome Trust (204829) through the Centre for Future Health (CFH) at the University of York, United Kingdom. The authors declare no conflicts of interest.
Figures

References
-
- Barrett F. F., McGehee R. F., Jr., Finland M. (1968) Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N. Engl. J. Med. 279, 441–448 - PubMed
-
- Udo E. E., Pearman J. W., Grubb W. B. (1993) Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J. Hosp. Infect. 25, 97–108 - PubMed
-
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., Liassine N., Bes M., Greenland T., Reverdy M. E., Etienne J. (2003) Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984 - PMC - PubMed
-
- Hiramatsu K., Aritaka N., Hanaki H., Kawasaki S., Hosoda Y., Hori S., Fukuchi Y., Kobayashi I. (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350, 1670–1673 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

