Identification and characterization of GmMYB118 responses to drought and salt stress
- PMID: 30509166
- PMCID: PMC6276260
- DOI: 10.1186/s12870-018-1551-7
Identification and characterization of GmMYB118 responses to drought and salt stress
Abstract
Background: Abiotic stress severely influences plant growth and development. MYB transcription factors (TFs), which compose one of the largest TF families, play an important role in abiotic stress responses.
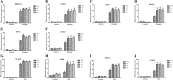
Result: We identified 139 soybean MYB-related genes; these genes were divided into six groups based on their conserved domain and were distributed among 20 chromosomes (Chrs). Quantitative real-time PCR (qRT-PCR) indicated that GmMYB118 highly responsive to drought, salt and high temperature stress; thus, this gene was selected for further analysis. Subcellular localization revealed that the GmMYB118 protein located in the nucleus. Ectopic expression (EX) of GmMYB118 increased tolerance to drought and salt stress and regulated the expression of several stress-associated genes in transgenic Arabidopsis plants. Similarly, GmMYB118-overexpressing (OE) soybean plants generated via Agrobacterium rhizogenes (A. rhizogenes)-mediated transformation of the hairy roots showed improved drought and salt tolerance. Furthermore, compared with the control (CK) plants, the clustered, regularly interspaced, short palindromic repeat (CRISPR)-transformed plants exhibited reduced drought and salt tolerance. The contents of proline and chlorophyll in the OE plants were significantly greater than those in the CK plants, whose contents were greater than those in the CRISPR plants under drought and salt stress conditions. In contrast, the reactive oxygen species (ROS) and malondialdehyde (MDA) contents were significantly lower in the OE plants than in the CK plants, whose contents were lower than those in the CRISPR plants under stress conditions.
Conclusions: These results indicated that GmMYB118 could improve tolerance to drought and salt stress by promoting expression of stress-associated genes and regulating osmotic and oxidizing substances to maintain cell homeostasis.
Keywords: CRISPR; Drought tolerance; Genome-wide analysis; MYB transcription factor; Salt tolerance; Soybean.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures









References
-
- Xu ZS, Chen M, Li LC, Ma YZ. Functions of the ERF transcription factor family in plants. Botany. 2008;86(9):969–977. doi: 10.1139/B08-041. - DOI
-
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

