Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells
- PMID: 30510202
- PMCID: PMC6277420
- DOI: 10.1038/s41598-018-35859-6
Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells
Abstract
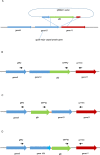
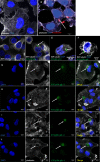
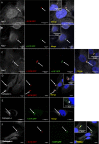
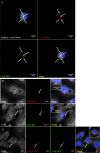
Bacterial infections can be treated with bacteriophages that show great specificity towards their bacterial host and can be genetically modified for different applications. However, whether and how bacteriophages can kill intracellular bacteria in human cells remains elusive. Here, using CRISPR/Cas selection, we have engineered a fluorescent bacteriophage specific for E. coli K1, a nosocomial pathogen responsible for urinary tract infections, neonatal meningitis and sepsis. By confocal and live microscopy, we show that engineered bacteriophages K1F-GFP and E. coli EV36-RFP bacteria displaying the K1 capsule, enter human cells via phagocytosis. Importantly, we show that bacteriophage K1F-GFP efficiently kills intracellular E. coli EV36-RFP in T24 human urinary bladder epithelial cells. Finally, we provide evidence that bacteria and bacteriophages are degraded by LC3-associated phagocytosis and xenophagy.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

