SumHer better estimates the SNP heritability of complex traits from summary statistics
- PMID: 30510236
- PMCID: PMC6485398
- DOI: 10.1038/s41588-018-0279-5
SumHer better estimates the SNP heritability of complex traits from summary statistics
Abstract
We present SumHer, software for estimating confounding bias, SNP heritability, enrichments of heritability and genetic correlations using summary statistics from genome-wide association studies. The key difference between SumHer and the existing software LD Score Regression (LDSC) is that SumHer allows the user to specify the heritability model. We apply SumHer to results from 24 large-scale association studies (average sample size 121,000) using our recommended heritability model. We show that these studies tended to substantially over-correct for confounding, and as a result the number of genome-wide significant loci was under-reported by about a quarter. We also estimate enrichments for 24 categories of SNPs defined by functional annotations. A previous study using LDSC reported that conserved regions were 13-fold enriched, and found a further six categories with above threefold enrichment. By contrast, our analysis using SumHer finds that none of the categories have enrichment above twofold. SumHer provides an improved understanding of the genetic architecture of complex traits, which enables more efficient analysis of future genetic data.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
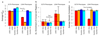
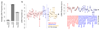

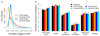
References
Publication types
MeSH terms
Grants and funding
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- U01 HG006382/HG/NHGRI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- U01 HG006380/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States
- RC4 AG039029/AG/NIA NIH HHS/United States
- U01 HG006385/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- RC2 AG036495/AG/NIA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MR/L012561/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

