Three-dimensional texture analysis of optical coherence tomography images of ovarian tissue
- PMID: 30511664
- PMCID: PMC6934175
- DOI: 10.1088/1361-6560/aaefd2
Three-dimensional texture analysis of optical coherence tomography images of ovarian tissue
Abstract
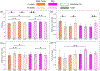
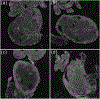
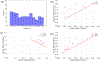
Ovarian cancer has the lowest survival rate among all gynecologic cancers due to predominantly late diagnosis. Optical coherence tomography (OCT) has been applied successfully to experimentally image the ovaries in vivo; however, a robust method for analysis is still required to provide quantitative diagnostic information. Recently, texture analysis has proved to be a useful tool for tissue characterization; unfortunately, existing work in the scope of OCT ovarian imaging is limited to only analyzing 2D sub-regions of the image data, discarding information encoded in the full image area, as well as in the depth dimension. Here we address these challenges by testing three implementations of texture analysis for the ability to classify tissue type. First, we test the traditional case of extracted 2D regions of interest; then we extend this to include the entire image area by segmenting the organ from the background. Finally, we conduct a full volumetric analysis of the image volume using 3D segmented data. For each case, we compute features based on the Grey-Level Co-occurence Matrix and also by introducing a new approach that evaluates the frequency distribution in the image by computing the energy density. We test these methods on a mouse model of ovarian cancer to differentiate between age, genotype, and treatment. The results show that the 3D application of texture analysis is most effective for differentiating tissue types, yielding an average classification accuracy of 78.6%. This is followed by the analysis in 2D with the segmented image volume, yielding an average accuracy of 71.5%. Both of these improve on the traditional approach of extracting square regions of interest, which yield an average classification accuracy of 67.7%. Thus, applying texture analysis in 3D with a fully segmented image volume is the most robust approach to quantitatively characterizing ovarian tissue.
Figures






References
-
- Abdolmanafi A, Duong L, Dahdah N, Cheriet F, 2017. Deep feature learning for automatic tissue classification of coronary artery using optical coherence tomography. Biomed. Opt. Express 8, 1203 URL: https://www.osapublishing.org/DirectPDFAccess/F77DD5AD-0BC7-36D6-5C4AA46..., doi:10.1364d/BOE.8.001203. - DOI - PMC - PubMed
-
- Abràmoff M, Garvin MK, Sonka M, 2010. Retinal imaging and image analysis. IEEE Rev. Biomed. Eng 1, 169–208. URL: http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=5660089, doi:10.1109/RBME.2010.2084567.Retinal. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
