Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways
- PMID: 30511668
- PMCID: PMC6277964
- DOI: 10.1107/S2053230X18015273
Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways
Abstract
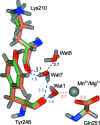
Lactobacillus brevis alcohol dehydrogenase (LbADH) is a well studied homotetrameric enzyme which catalyzes the enantioselective reduction of prochiral ketones to the corresponding secondary alcohols. LbADH is stable and enzymatically active at elevated temperatures and accepts a broad range of substrates, making it a valuable tool in industrial biocatalysis. Here, the expression, purification and crystallization of LbADH to generate large, single crystals with a volume of up to 1 mm3 suitable for neutron diffraction studies are described. Neutron diffraction data were collected from an H/D-exchanged LbADH crystal using the BIODIFF instrument at the Heinz Maier-Leibnitz Zentrum (MLZ), Garching, Germany to a resolution dmin of 2.15 Å in 16 days. This allowed the first neutron crystal structure of LbADH to be determined. The neutron structure revealed new details of the hydrogen-bonding network originating from the ion-binding site of LbADH and provided new insights into the reasons why divalent magnesium (Mg2+) or manganese (Mn2+) ions are necessary for its activity. X-ray diffraction data were obtained from the same crystal at the European Synchrotron Radiation Facility (ESRF), Grenoble, France to a resolution dmin of 1.48 Å. The high-resolution X-ray structure suggested partial occupancy of Mn2+ and Mg2+ at the ion-binding site. This is supported by the different binding affinity of Mn2+ and Mg2+ to the tetrameric structure calculated via free-energy molecular-dynamics simulations.
Keywords: Lactobacillus brevis; alcohol dehydrogenase; hydrogen-bonding network; neutron diffraction; protein crystallization; short-chain dehydrogenases/reductases.
Figures







References
-
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. - PMC - PubMed
-
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Echols, N., Headd, J. J., Hung, L. W., Jain, S., Kapral, G. J., Grosse Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2011). Methods, 55, 94–106. - PMC - PubMed
-
- Allnér, O., Nilsson, L. & Villa, A. (2012). J. Chem. Theory Comput. 8, 1493–1502. - PubMed
-
- Bennett, C. H. (1976). J. Comput. Phys. 22, 245–268.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

